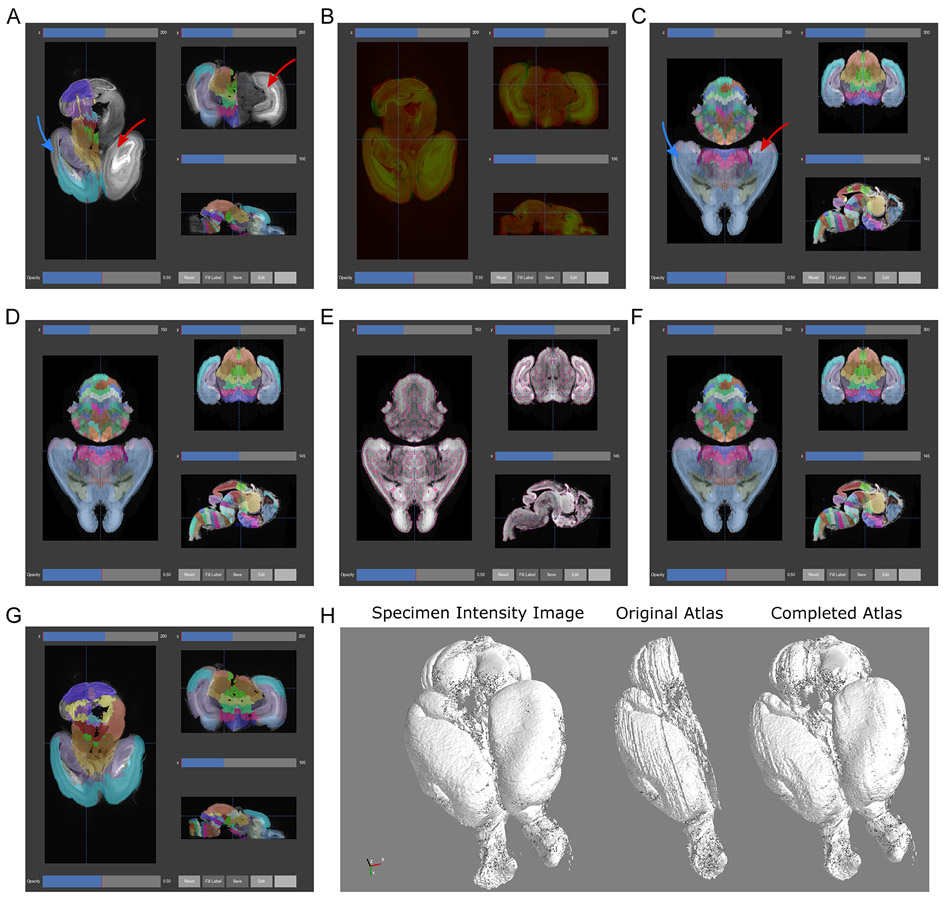
Figure 7.
Atlas registration and automated refinement. (A) Registration of the original Allen Developing Mouse Brain Atlas E18.5 atlas to the sample P0 mouse brain. After registration, the existing labels approximate the position, orientation, size, and shape of the corresponding parts of the sample brain. Note the missing labels in one entire hemisphere (red arrow) and the lateral planes of the labeled hemisphere (blue arrow). (B) Overlay of the sample brain (green) and registered atlas (red) intensity images as a qualitative assessment of registration. Misalignments can be optimized by adjusting settings in the software’s atlas profiles. (C) The automated 3D atlas generation pipeline performs extension of lateral labels into the unlabeled areas (blue arrow) by iteratively growing the existing labels to fit the anatomical contours (part F). After extending the lateral labels, the unlabeled hemisphere is replaced by mirroring the labeled hemisphere (red arrow), resulting in a fully labeled atlas in 3D. (D) Label smoothing removed many of the jagged artifacts most visible in the axial and coronal planes. (E) To further smooth and refine labels, edge detection (pink lines) of gross anatomical boundaries (high grayscale contrast) in the microscopy intensity image provides a 3D guide for aligning label edges. (F) Starting with the atlas from part C, each label is eroded to its core and regrown by a watershed algorithm guided by the anatomical map from part E, followed by smoothing as in part D to remove small artifacts from the watershed. (G) The same registration as performed in part A is performed but with the generated 3D atlas labels. (H) The 3D Viewer provides a macroscopic view of the specimen through 3D surface rendering of the sample brain (left). Surface rendering of the registered labels (center) before and (right) after 3D atlas generation demonstrate label completion and smoothing.