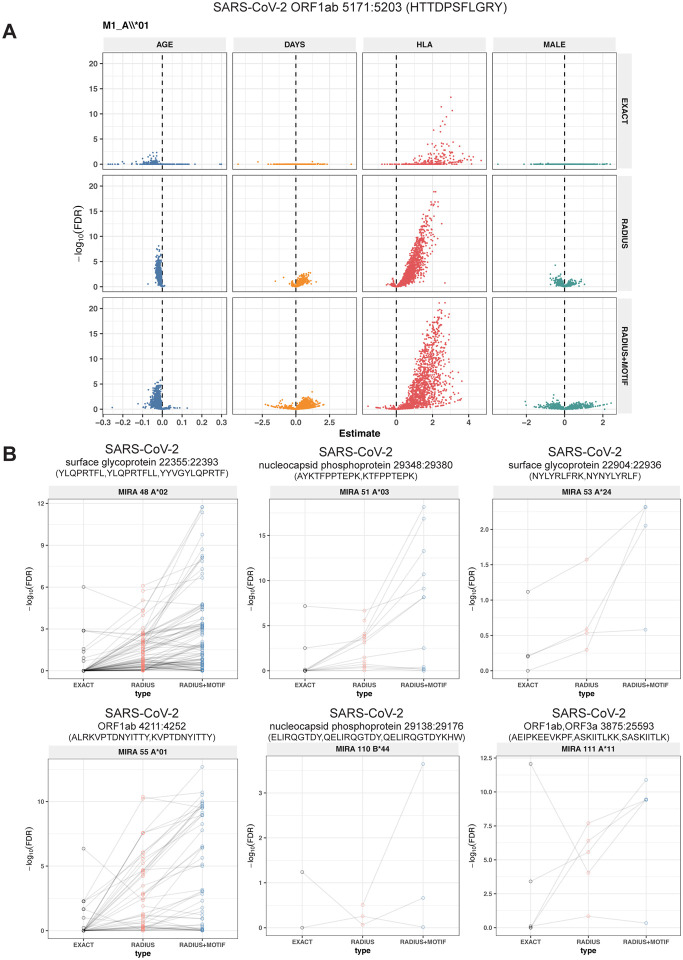
Figure 6. Associations of TCR features with participant age, days post diagnosis, HLA-genotype, and sex in TCR β-chain repertoires of COVID-19 patients (n=694).
(A) Beta-binomial regression coefficient estimates (x-axis) and negative log10 false discovery rates (y-axis) for features developed from CD8+ TCRs activated by SARS-CoV-2 MIRA55: ORF1ab 5171:5203. The abundances of TCR meta-clonotypes are more robustly associated with predicted HLA type than exact clonotypes. (B) Signal strength of enrichment by participant HLA-type (2-digit) of TCR β-chain clonotypes (EXACT) and meta-clonotypes (RADIUS or RADIUS+MOTIF) predicted to recognize additional HLA-restricted SARS-CoV-2 peptides: (1) MIRA48: surface glycoprotein 22355:22393 (2) MIRA51: nucleocapsid phosphoprotein 29348:29380), (3) MIRA53: surface glycoprotein 22904:22936 (4) MIRA55: ORF1ab 4211:4252 (5) MIRA110: nucleocapsid phosphoprotein 29138:29176 (6) MIRA11: ORF1ab, ORF3a 3875:25593. Models were estimated with counts of TCR matching clonotypes (EXACT) or meta-clonotypes (RADIUS or RADIUS+MOTIF) with the following definitions: (1) EXACT (inclusion of TCRs matching the centroid TRBV gene and CDR3 amino acids), (2) RADIUS (TCR centroid with inclusion criteria defined by an optimized TCRdist radius), (3) RADIUS + MOTIF (inclusion criteria defined by TCR centroid, optimized radius and matching of CDR3 amino acid residues at conserved positions within the meta-clonotype CDR3s). see Methods for details.