Figure 2. Analysis of 23 Patient derived whole SARS-CoV-2 genome sequences in context of national sequences and other cases of chronic SARS-CoV-2 shedding.
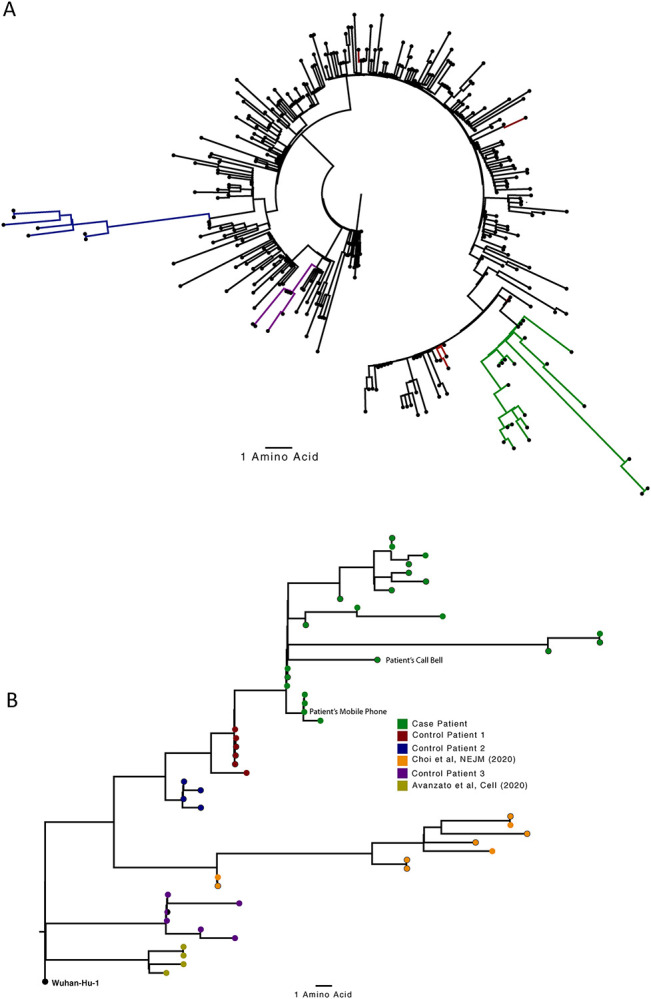
A. Circularised maximum-likelihood phylogenetic tree rooted on the Wuhan-Hu-1 reference sequence, showing a subset of 250 local SARS-CoV-2 genomes from GISAID. This diagram highlights significant diversity of the case patient (green) compared to three other local patients with prolonged shedding (blue, red and purple sequences). All “United Kingdom / English” SARS-CoV-2 genomes were downloaded from the GISAID database and a random subset of 250 selected as background. B. Close-view maximum-likelihood phylogenetic tree indicating the diversity of the case patient and three other long-term shedders from the local area (red, blue and purple), compared to recently published sequences from Choi et al (orange) and Avanzato et al (gold). Control patients generally showed limited diversity temporally, though the Choi et al sequences were found to be even more divergent than the case patient. Environmental samples (patient’s call bell, and patient’s mobile phone) are indicated.
