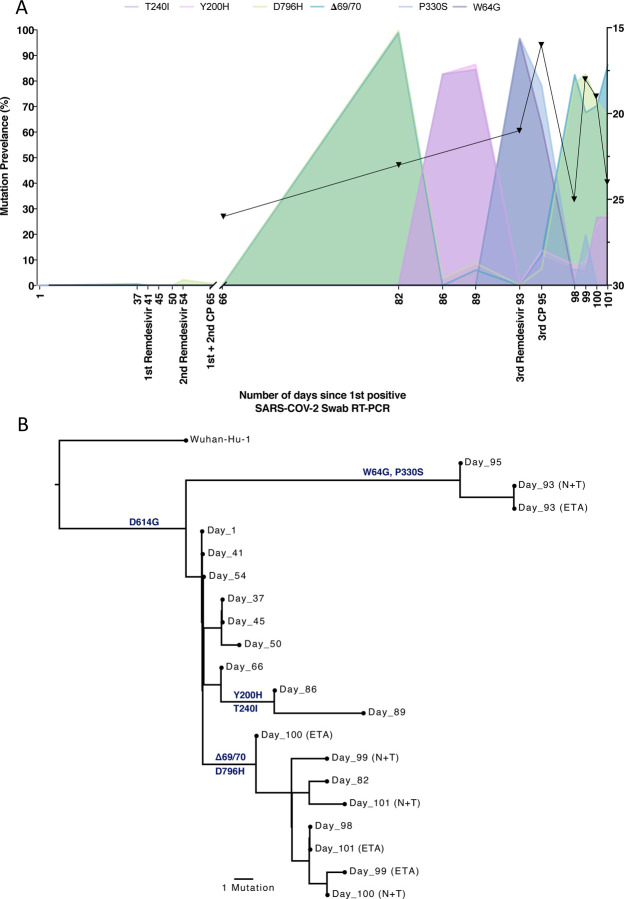
Figure 5. Longitudinal variant frequencies and phylogenetic relationships for virus populations bearing six Spike (S) mutations.
A. At baseline, all four S variants (Illumina sequencing) were absent (<1% and <20 reads). Approximately two weeks after receiving two units of convalescent plasma (CP), viral populations carrying ΔH69/V70 and D796H mutants rose to frequencies >90% but decreased significantly four days later. This population was replaced by a population bearing Y200H and T240I, detected in two samples over a period of 6 days. These viral populations were then replaced by virus carrying W64G and P330S mutations in Spike, which both dominated at day 93. Following a 3rd course of remdesivir and an additional unit of convalescent plasma, the ΔH69/V70 and D796H virus population re-emerged to become the dominant viral strain reaching variant frequencies of >90%. Pairs of mutations arose and disappeared simultaneously indicating linkage on the same viral haplotype. CT values from respiratory samples are indicated on the right y-axis (black triangles) B. Maximum likelihood phylogenetic tree of the case patient with day of sampling indicated. Spike mutations defining each of the clades are shown ancestrally on the branches on which they arose. On dates where multiple samples were collect, these are indicated as endotracheal aspirate (ETA) and Nose + throat swabs (N+T).