Abstract
The manuscript presents a bragging-based ensemble forecasting model for predicting the number of incidences of a disease based on past occurrences. The objectives of this research work are to enhance accuracy, reduce overfitting, and handle overdrift; the proposed model has shown promising results in terms of error metrics. The collated dataset of the diseases is collected from the official government site of Hong Kong from the year 2010 to 2019. The preprocessing is done using log transformation and z score transformation. The proposed ensemble model is applied, and its applicability to a specific disease dataset is presented. The proposed ensemble model is compared against the ensemble models, namely dynamic ensemble for time series, arbitrated dynamic ensemble, and random forest using different error metrics. The proposed model shows the reduced value of MAE (mean average error) by 27.18%, 3.07%, 11.58%, 13.46% for tuberculosis, dengue, food poisoning, and chickenpox, respectively. The comparison drawn between the proposed model and the existing models shows that the proposed ensemble model gives better accuracy in the case of all the four-disease datasets.
Keywords: Bootstrapping, Bragging, Disease forecasting, Ensemble, Time series forecasting
Introduction
The rising prevalence of data acquisition applications leads to the collection of a vast amount of time-series data that enable forecasting for many medical applications. A multitude of end-use cases for time-series applications in the medical domain exists that aims to process thousands of time-series and millions of data points on an immense scale. Across the world, human populations are afflicted by numerous ill-health conditions. Several of these diseases are highly contagious and, therefore, reliable and sustainable predictions in the clinical setting need to be focused upon. These predictions are based on the available data regarding its spread across the physical and geographical spectrum. The presented research work focuses on such ill-health manifestations worldwide and determines ways of predicting their cyclical occurrences with a high degree of reliability. A bragging based heterogeneous ensemble model for predicting the future incidences of the diseases is proposed. The diseases selected in this research work are dengue, tuberculosis, chickenpox, and food poisoning. The number of incidences of these diseases is significant in the field of epidemiology. Also, these diseases show different trend patterns enabling them to evaluate the proposed ensemble model more extensively.
Dengue is the most prevalent mosquito-borne infection that has increased 30-fold in the past 5 decades and is declared an endemic in over 100 countries having around 3.9 billion people are at the risk of dengue infection [1]. Around 2.7 million cases and 1206 deaths in the year 2019 in America have been reported by the PAHO (Pan American Health Organization) [2], and out of the total risks spreading in 129 countries, 70% of the infections are in Asia [3]. Further, TB (tuberculosis), a highly infectious disease caused by the bacillus mycobacteria, is a major cause of sickness and is one of the ten reasons for death worldwide [4]. The number of TB cases varies from under 5 to above 500 new incidences per 100,000 population, with the average being 130 worldwide [5]. As reported by WHO (World Health Organization), 22 countries are in the list of 30 profoundly affected countries, with a high mortality rate, an estimated 3.7 million deaths reported in the year 2018. Correspondingly, the disease affects both genders, with males accounting for 57% cases, females 32%, and children being 11% of the total affected population [6]. Further, chickenpox outbreaks keep on happening even in those settings where most kids are inoculated, for example, schools [7]. The medical data collected at CDC (Centers for Disease Control and Prevention) claim a substantial decline regarding chickenpox outpatient visits and hospitalization by 84% and 93%, respectively, as compared to the pre-vaccination period. The mortality rate due to the disease also reduced to 87% compared to the pre-vaccine year [8]. Likewise, FP (food poisoning), a prevalent worldwide disease, occurs because of the consumption of infected or polluted food [9]. WHO estimates that 600 million people, i.e., 1 out of 10 people fall ill due to food poisoning, and 4,20,000 expire yearly. Out of these, 40% of the disease burden is carried by the children below 5 years of age [10]. Despite the continuous efforts put by leading public health organizations, the spread of viral diseases remains high across the globe. If not restricted before-hand, it can lead to the deployment of extra resources (human, infrastructure, and financial resources) and may bring the whole society at a standstill [11]. Failure to handle pandemic diseases in an effective manner may result in high morbidity and mortality [12]. Therefore, a robust health system mechanism and preparedness for the detection, confirmation, and control of such diseases are of utmost importance. The manuscript presents a novel model to predict the number of incidences of the diseases and evaluates its efficacy by comparing it with the state-of-the-art models. The proposed research work’s novelty lies in an ensemble model that takes seasonal dependency into account and model trend and seasonality separately.
Objective
The main objective of the research work presented here is to propose a heterogeneous ensemble model to handle the three main drawbacks of ensembling, i.e., concept drift, overfitting, and noise. The proposed model is designed to handle the seasonality and trend separately. For trend modeling, an ensemble of ARIMA (Auto-Regressive Integrated Moving Average), SNaive, Spline, and NNAR (Neural Network Auto Regression) is applied. Modeling of Seasonality is done using CART (Classification and Regression Trees). Bootstrapping of CART is done to avoid overfitting, and the median of all the forecasts is taken as the final forecast. Median is less prone to outliers and is not senstive to large fluctuations as compared to the mean because significant variations do not impact the median much [13]. Hence, the proposed ensemble overcomes the drawbacks of ensembling and helps in achieving higher prediction accuracy.
Significance
Time series forecasting involves the application of statistical models to predict future incidences based on past data. These models, when applied independently, can fit the data and generate accurate forecasts. However, these models suffer from a drawback of poorly adapting to unexpected conditions, resulting from some environmental factors or particular sudden virus spread situations [14]. In scientific literature, this condition is known as “concept drift”. Modeling of concept drift implies finding the unknown or hidden relationship between input and output variables. Also, in disease forecasting, time-series data are not always stationary [15], and thus the distribution of data, i.e., the concept changes unexpectedly with time. Therefore, it becomes imperative to consider the concept of concept drift while forecasting. Further, ensemble forecasting techniques are universally applied as de facto standard nowadays for predictive accuracy improvement. This involves combining multiple predictions from base models to enhance the accuracy of base models and avoid overfitting. The significance of the presented research work is to propose a bragging based ensemble model for disease forecasting and to check its efficacy on the disease datasets.
Background
A brief review of the various methodologies in disease forecasting and modeling techniques can generate new insights into the field of disease modeling. Preliminary work in this field focused mostly on the application of time series forecasting models for disease forecasting. A seminal work done by the International Society for Disease Surveillance proposed a framework for disease outbreak detection and validated the framework by comparing it with several variants and evaluated forecast residuals to alert the concerned authorities [16]. An ensemble learning paradigm for FCM (fuzzy cognitive map) is proposed for the classification of Autism, and the results are significantly better in terms of accuracy [17].
Besides, Smith et al. proposed a multi-model ensemble framework for predicting transmission and elimination dynamics of lymphatic filariasis. This multi-model ensemble framework overcomes the biases of a single model [18]. Yin and Jha employed machine learning ensemble models based on wearable medical sensors and discovered that the third prominent cause of death in a developed country like the US is preventable medical negligence [19]. This negligent behavior is avoided by a CDSSs (computer-based clinical decision support systems). Furthermore, the potential and efficiency of ensemble models are also witnessed in [20]. The researchers here worked on the spread of the Ebola virus from 2014 to 2015 and established that the ensemble model significantly improves prediction accuracy over any individual modeling approach.
Shaman and Karspeck proposed the first step towards developing a statistical system for influenza forecasting and developed a framework for forecasting seasonal outbreaks of influenza using data assimilation technique and can predict seven days ahead [21]. Zarebski et al. used PF (particle filters) in association with mechanistic transmission models to forecast seasonal influenza progression and predicted the count of laboratory-confirmed occurrences of influenza in Melbourne in 2010–2015 epidemics [22].
Further, Cobb et al. made a comparison of two Bayesian data assimilation techniques for tracking the COVID-19 epidemic [23]. The simulations were performed for tracking the spatio-temporal patterns of emerging epidemics. The parameter estimation of TB, a prime cause of mortality across the globe, is undertaken by authors in [24]. Here, the authors employed the EnKF (ensemble Kalman filter) approach to estimate the smear-positive cases in India for the period 2006–2011. Kalman filter has also been employed to predict various infectious diseases like HIV/AIDS, measles, and influenza [25].
Additionally, ensemble modeling has proven to outperform existing base models in other domains as well. Sainan et al. [26] conducted a study on the bagging method and found that the bagging method outperforms the historical mean method when the in-sample estimation period is small. When the number of observations for in-sample is 24, 60, and 120, the percentage gains of the bagging method over the historical mean in terms of out-of-sample are around 0.45–4.6% and − 2.8 to 0.7% positive and negative, respectively. Fang et al. [27] compared the RF (random forest) model with autoregressive integrated moving average ARIMA models using morbidity and meteorological data from 2012 to 2016 to train the models and data from 2017 for testing purposes. RF model extensively combined the simultaneous and lagged outcomes of meteorological elements; it additionally incorporated the autocorrelation and irregularity of the morbidity. Further, Rathore et al. proposed an ensemble model to predict faults in software modules [28, 29]. Here, authors employ a combination of approaches based on linear and non-linear combination rule for the ensemble. The results prove the efficacy of ensembling in this field as well. Similarly, Rathore et al. presented a dynamic approach to select the base learners for software fault prediction [30]. In this seminal work, initially, the neighbor module the same as the testing module is located based on a distance function, and then the best learner is selected. The method demonstrated a remarkable predictive accuracy enhancement.
As observed that ensemble modeling approaches have been applied to forecast infectious diseases. It can be safely concluded that most of the studies have focused on enhancing the prediction accuracy using ensembling techniques. Nevertheless, the concept of "concept drift" remains uncharted in the research domain. In epidemiology, the data collected at different time points are not always stationary; in other words, seasonality changes unexpectedly with time. Therefore, it becomes imperative to incorporate seasonal dependency into account.
Proposed Bragging-Based Heterogeneous Ensemble Model for Disease Forecasting
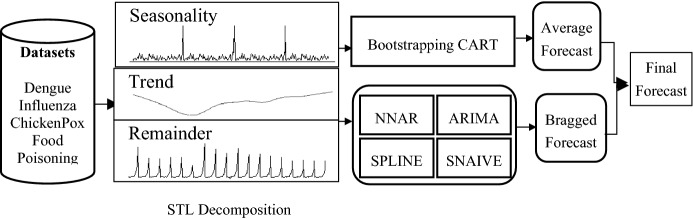
This section presents the heterogeneous bragging-based ensemble model for disease forecasting. Data pertaining to diseases are known to have strong seasonal dependencies; hence, the proposed model integrates the dependencies exhibited in the data. Depending on seasonal dependencies, forecasts will be computed monthly, quarterly, or yearly. The working methodology is demonstrated in Fig. 1; The dataset is decomposed into seasonality, trend, and a residual component. The trend component is modeled using an ensemble of base models, namely ARIMA, SNaive, Spline, and NNAR. The final forecast of this ensemble is the average of the forecast of the base models. A bootstrap sample with replacement is created for the seasonality component, and CART is applied as a bragged ensemble model. The output of the model is taken as the median of all bootstrapped forecasts. Median is less prone to outliers, therefore, selected to combine the forecasts. The final forecast is the aggregated of the forecast of the seasonality component and trend component.
Fig. 1.
Proposed bragging-based ensemble model demonstrating the modeling of trend using ensemble model and seasonality modeling using bragged ensemble model
The algorithm for the proposed methodology is as follows:
- The prediction of no. of cases () this year on the basis of past measurements (, ,…,) can be represented using linear coefficient (), (, ,…,) and error coefficient (). Autocorrelation function is applied to determine the coefficient of the time series.
1 - The Fourier transform is done to get the weights of the components of the time-series.
2 - The series is checked for stationarity by performing ADF (augmented Dickey–Fuller) test on the series. If the series is nonstationary, differencing is done to convert the series into stationary form. Mathematically, it is represented as
3 An additive STL (seasonality trend estimation using loess) decomposition is performed on the time series data to divide it into its three components, i.e. seasonality , trend , and residual . Additive STL decomposition is suited the most appropriate when the seasonality does not vary as the variation in the time series. If the seasonal variations are more, multiplicative STL decomposition is performed. Residual component is removed from the decomposition.
- Base models are applied to the trend part of the decomposed time series. The final value of trend forecast is calculated as the average of all the models.
4 - Seasonality is sampled times randomly and constructing a bootstrap Sample (, ……, ) with replacement from the values (). Next is to compute the bootstrapped forecast as
5 - Step 6 is repeated no. of times to create the final seasonality forecast is obtained by taking the median of all the forecasts.
6 Final forecast is evaluated by aggregating trend forecast and seasonality forecast.
The pseudocode of the proposed model is given below.
Materials and Methods
This section provides the details of applying the proposed data ensemble technique on the selected dataset and stepwise experiments performed on the proposed technique. The statistical tool termed R is employed for the application of the proposed model and disease forecasting. Experimental disease data come from the Center for Health Protection, Department of Health (https://www.chp.gov.hk). Monthly Chickenpox disease data give the total number of infections from the year 1999 to 2019. In contrast, Food Poisoning data are from the year 2000 to 2019 and contains the reason for the food poisoning like bacteria, chemicals, viruses, biotoxin, and other factors. Dengue data are collected from the year 2002 to 2019. Tuberculosis data are from the year 1995 to 2019. The preprocessing, i.e., cleaning, handling missing values by replacing with the average value, is applied to the disease data, and the data are prepared for further processing. Further, the data split is done into the training and testing set in the ratio of 80% and 20%, respectively. Table 1 represents the sample of the tuberculosis dataset collected for experimentation purpose. The dataset represents the no. of cases and deaths of male and female patients. A comprehensive description of the methods employed in the model is given below.
Table 1.
Statistics measures of ADF test
| Year | Month | Outbreaks | Persons affected |
|---|---|---|---|
| 2019 | January | 7 | 26 |
| 2019 | February | 24 | 216 |
| 2019 | March | 21 | 90 |
| 2019 | April | 17 | 62 |
| 2019 | May | 24 | 74 |
| 2019 | June | 35 | 90 |
| 2019 | July | 20 | 86 |
| 2019 | August | 11 | 26 |
| 2019 | September | 21 | 60 |
| 2019 | October | 17 | 69 |
| 2019 | November | 5 | 12 |
| 2019 | December | 10 | 37 |
Handling Seasonality
CART is a recursive tree-based method that finds the best fit by selecting the node which maximizes the information gain. RPART implementation of this method in R is used to create binary trees. Lag and cosine of the daily and weekly season are found to be the variables of most importance. The number of splits is taken to be 600. Hyperparameter tuning is done manually by keeping the complexity parameter as 0.000001, min split to 2, and max depth to 30.
Handling Trend
ARIMA() implementation of the ARIMA forecast in R is applied by keeping the confidence level as 95. NNAR handles the seasonality as ARIMA, but it also handles the nonlinear functions. NNAR is implemented through the method nnetar() available in the forecast package of R. SNaive is the method of handling high seasonal data, and is implemented using R implementation as snaive() by keeping the value as 5. Spline is a non-parametric function that is used to smooth the data by interpolation. The implementation used the Spline() implementation of R.
Handling Overfitting
Overfitting occurs when the fitted model does not generalize well to the unseen data. This can be calculated by calculating the predictive accuracy of the model on the training data as well as testing data. For handling overfitting, 100 values of bootstrapped forecasts by CTREE (Conditional Inference Trees) are performed. Sampling for the bootstrapping is done with replacement in the ratio of 70%. RPART (Recursive Partitioning And Regression Trees) value of MAPE (Mean Absolute Percentage Error) metrics of tuberculosis, dengue data, food poisoning, and chickenpox, when applied on the training data, comes out to be 0.55, 0.01, 0.28, 0.36, respectively, which shows that the model fits accurately on the training data. However, in the case of testing data, the MAPE error metrics of tuberculosis, dengue data, food poisoning, and chickenpox are 32.43, 35.6, 43.12, 56.71, respectively. The testing error metrics clearly shows the problem of overfitting. Hence, bootstrapping is going to be a viable solution.
Time Series Representation and STL Decomposition
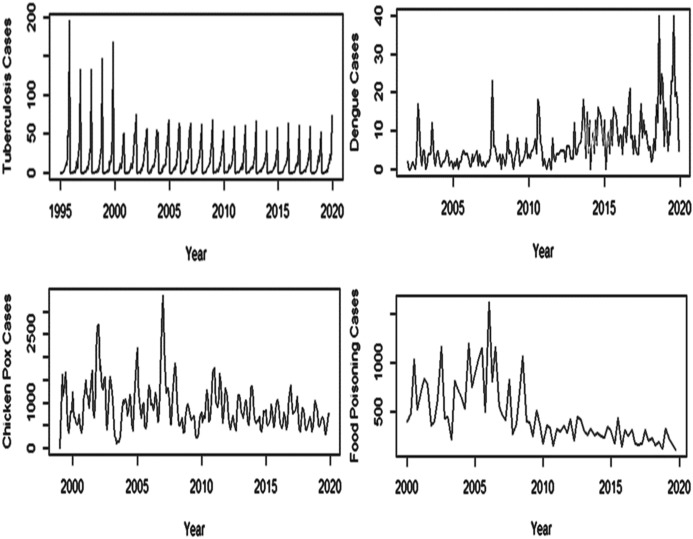
Time series representation is used to represent the collected data, training data, and test data, respectively, as demonstrated in Fig. 2. The graph in the series represents the original time series representation of the data demonstrating trend and seasonality. To understand the three essential properties, namely seasonality, trend, and residual, STL decomposition (based on loess regression) of the time series data is applied. Out of the four selected diseases, the trend of TB is almost constant; dengue cases show an upward trend, whereas chickenpox and food poisoning show a declining trend. TB dataset shows a concept drift in 1995 and 2000; dengue in the year 2018 and 2019; chickenpox in 2007; and food poisoning in the year 2006.
Fig. 2.
Time series representation of disease datasets to demonstrate the trend and seasonality in the data
Application of Ensembling Model
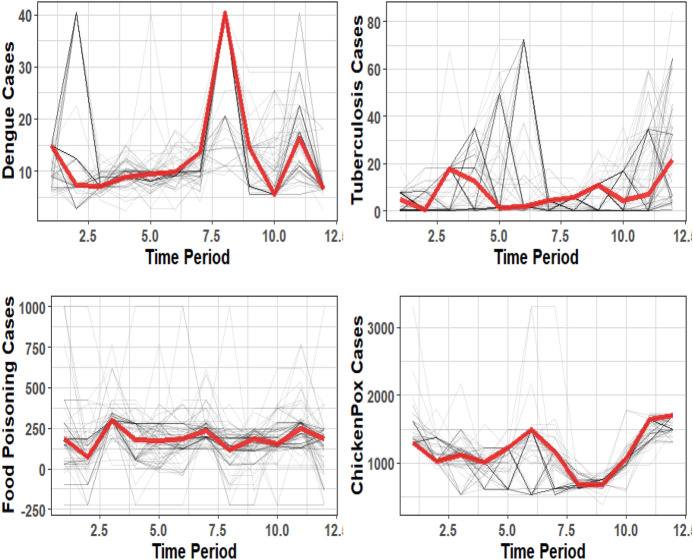
The training dataset is sampled with replacement with a ratio of 70%. The models are trained on a new train set. 100 boots are taken, and the procedure is repeated for 100 boots. The median of the predictions is taken as the final aggregate prediction. The training set consists of the 1-year lagged data, and the double seasonality Fourier series is applied. In Fig. 3 are the graphs of the median of the forecasts.
Fig. 3.
Application of ensemble model on the bootstrapped datasets. Lighter grey lines represent the individual forecasts of the model. Red line represents the final forecast
Results and Discussion
To check the stationarity of the time series data, the ADF test is done. The value of the test statistics is given in Table 2.
Table 2.
Statistics measures of ADF test
| 0.815369 | |
| 0.991880 | |
| 13.000000 | |
| − 3.481682 | |
| − 2.884042 | |
| − 2.578770 |
If the test statistics’ value is greater than the critical value, the time series is non-stationary. The results demonstrated in Table 1 show that the statistics is higher than the critical value, and hence the series is not stationary. The coefficient of determination () is used to determine the coefficient of time series.
Table 3 comprises average errors of predictions as different error metrics for base models and the ensemble model. The ensemble model is found to be the most accurate where the error metrics are minimum for all the disease datasets. Also, the datasets of chickenpox and food poisoning, which uncover higher values of error metrics for base models, have also shown a significant decline in the values of error metrics. MAE, i.e., the mean average error of the ensemble model for the tuberculosis, dengue, food poisoning, and chickenpox are 1.58, 1.89, 11.45, and 15.68, respectively. Amongst the base models, NNAR has shown the best performance with the MAE error metrics being: 2.97, 2.75, 14.59, and 24.12 for diseases in the same order as given above, respectively. The ensemble model enhances the predictive accuracy for the tuberculosis, dengue, food poisoning, and chickenpox by 46.80%, 31.27%, 21.52%, 34.99% as compared to the independent NNAR model, respectively.
Table 3.
MAE error metrics result after the application of base model on disease datasets
| Base learners | MAE | |||
|---|---|---|---|---|
| Tuberculosis | Dengue | Food poisoning | Chicken pox | |
| Theta forecasting | 13.40 | 2.48 | 49.65 | 54.12 |
| Moving average | 14.63 | 4.58 | 56.01 | 46.86 |
| Spline | 13.64 | 4.19 | 37.06 | 33.71 |
| Naïve | 9.56 | 3.84 | 21.58 | 30.41 |
| Random walk with drift | 9.55 | 3.84 | 42.25 | 44.25 |
| Croston’s method | 16.58 | 3.31 | 49.96 | 47.97 |
| Holt winter | 12.57 | 3.36 | 47.58 | 44.41 |
| Simple exponential smoothing | 12.55 | 3.34 | 65.38 | 43.44 |
| Seasonal naïve | 3.59 | 3.43 | 18.57 | 39.24 |
| NNAR | 2.97 | 2.75 | 14.59 | 24.12 |
| ETS | 4.26 | 2.88 | 88.31 | 56.46 |
| ARIMA | 3.50 | 2.92 | 31.78 | 37.78 |
Further, the base models selected for modeling the trends of the dataset also have shown the minimum value of the error metrics, which further establishes the justification of the selected base models. NNAR, ARIMA, SNaive, Spline demonstrates the lowest value of error metrics with NNAR outperforming all the base models with values being 2.97, 2.75, 14.59, and 24.12 for tuberculosis, dengue, food poisoning, and chickenpox, respectively. The value of different error metrics for the NNAR base model is given in Table 3. The value evaluated is 0.89, representing a good fit. The Wilcoxon rank sum test [19] is applied to evaluate the performance of the proposed model with respect to the base models. As the residual errors are non-parametric, so a non-parametric test is applied to check the validity of the proposed model. A significance level α = 0.05 showed that the proposed model performs better than the base models (Table 4).
Table 4.
Error metrics result of best performing base model (NNAR) on disease datasets
| Disease | RMSE | MAE | MAPE |
|---|---|---|---|
| Tuberculosis | 4.34 | 2.97 | 0.82 |
| Dengue | 3.70 | 2.75 | 0.82 |
| Food poisoning | 19.57 | 14.59 | 0.76 |
| Chickenpox | 13.50 | 24.10 | 0.39 |
Further, state of the art ensemble models, namely DETS (Dynamic Ensemble for Time Series) [31], ADE (Arbitrated Dynamic Ensemble) [32], and Random Forest [33] are implemented, and a comparison is made with the proposed ensemble model in Table 5.
Table 5.
Error metrics result after the application of proposed model on disease datasets
| Ensemble models | RMSE | MAE | MAPE |
|---|---|---|---|
| DETS-time series ensemble model (tuberculosis) | 4.15# | 2.17# | 0.86# |
| ADE-time series ensemble model (tuberculosis) | 4.27 | 2.56 | 0.87 |
| Random forest (tuberculosis) | 5.86 | 3.04 | 1.24 |
| Proposed ensemble model (tuberculosis) | 2.25# | 1.58# | 0.54# |
| DETS-time series ensemble model (dengue) | 3.19# | 1.95# | 0.78# |
| ADE-time series ensemble model (dengue) | 3.26 | 2.31 | 0.91 |
| Random forest (dengue) | 5.26 | 3.56 | 1.13 |
| Proposed ensemble model (dengue) | 1.43# | 1.89# | 0.34# |
| DETS-time series ensemble model (food poisoning) | 14.51 | 11.95 | 0.25 |
| ADE-time series ensemble model (food poisoning) | 16.23 | 12.34 | 0.31 |
| Random forest (food poisoning) | 19.54 | 21.87 | 0.95 |
| Proposed ensemble model (food poisoning) | 11.56# | 11.45# | 0.13# |
| DETS-time series ensemble model (chickenpox) | 14.53 | 18.12 | 0.21 |
| ADE-time series ensemble model (chickenpox) | 15.31 | 20.12 | 0.28 |
| Random forest (chickenpox) | 20.08 | 24.53 | 0.34 |
| Proposed ensemble model (food poisoning) | 12.78# | 15.68# | 0.26# |
# indicates minimum value of the error metric
The proposed ensemble model is compared with the best performing existing ensemble model and the percentage comparison is given in Table 6. The results show the efficacy of the proposed model with existing state-of-the-art ensemble models.
Table 6.
Comparison of percentage decrease in error metrics of proposed ensemble model vis-à-vis other ensemble models
| Ensemble models | RMSE (%) | MAE (%) | MAPE (%) |
|---|---|---|---|
| Proposed ensemble model (tuberculosis) | 45.17 | 27.18 | 37.20 |
| Proposed ensemble model (dengue) | 55.17 | 3.07 | 56.41 |
| Proposed ensemble model (food poisoning) | 20.33 | 4.18 | 48 |
| Proposed ensemble model (food poisoning) | 12.04 | 13.46 | − 23.8 |
Conclusions
In summary, the research work exhibits the application of the ensembling approach to blend forecasts from multiple models of disease incidence prediction. The work presented here, along with predictive accuracy enhancement, focuses on three main properties, precisely concept drift, overfitting, and handling outliers. The proposed model shows promising results in terms of error metrics when compared with state-of-the-art ensemble models. As more data related to communicable disease outbreaks are operationalized and merged into public health data repositories, it will be more significant to combine dissimilar forecasts and merge information from each so as to obtain the most accurate prediction of an unfolding disease outbreak. For future scope, more geographical and temporal dependencies can be incorporated to make the model more robust concerning regional outbreaks.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nonita Sharma, Email: nonita@nitj.ac.in.
Jaiditya Dev, Email: jaidityadev1402@gmail.com.
Monika Mangla, Email: manglamona@gmail.com.
Vaishali Mehta Wadhwa, Email: wadhwavaishali@gmail.com.
Sachi Nandan Mohanty, Email: sachinandan09@gmail.com.
Deepti Kakkar, Email: kakkard@nitj.ac.in.
References
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allicock OM, Lemey P, Tatem AJ, Pybus OG, Bennett SN, Mueller BA, Suchard MA, Foster JE, Rambaut A, Carrington CVF. Phylogeography and population dynamics of dengue viruses in the Americas. Mol. Biol. Evol. 2012;29:1533–1543. doi: 10.1093/molbev/msr320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 2012;6(8):1–15. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glaziou P, Floyd K, Raviglione MC. Global epidemiology of tuberculosis. Semin Respir Crit Care Med. 2018;39(03):271–285. doi: 10.1055/s-0038-1651492. [DOI] [PubMed] [Google Scholar]
- 5.MacNeil A, Glaziou P, Sismanidis C, Maloney S, Floyd K. Global epidemiology of tuberculosis and progress toward achieving global targets—2017. Morb. Mortal. Wkly. Rep. 2019;68:263. doi: 10.15585/mmwr.mm6811a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wharton M. The epidemiology of varicella-zoster virus infections. Infect. Dis. Clin. 1996;10:571–581. doi: 10.1016/S0891-5520(05)70313-5. [DOI] [PubMed] [Google Scholar]
- 7.Lopez AS, LaClair B, Buttery V, Zhang Y, Rosen J, Taggert E, Robinson S, Davis M, Waters C, Thomas CA, et al. Varicella outbreak surveillance in schools in sentinel jurisdictions, 2012–2015. J. Pediatr. Infect. Dis. Soc. 2019;8:122–127. doi: 10.1093/jpids/piy010. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (CDC) Evolution of varicella surveillance–selected states, 2000–2010. MMWR Morb. Mortal. Wkly. Rep. 2012;61(32):609. [PubMed] [Google Scholar]
- 9.Richa S, Vijay S, Ruchi S, Prakash GO. Etiological and clinical characteristics of a diarrhea epidemic in Western Indian State. Clin. Gastroenterol. Hepatol. 2017;15(1):e27–e28. [Google Scholar]
- 10.Tripathi NK, Shrivastava A. Recent developments in recombinant protein–based dengue vaccines. Front. Immunol. 2018;9:1919. doi: 10.3389/fimmu.2018.01919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain R, Sontisirikit S, Iamsirithaworn S, Prendinger H. Prediction of dengue outbreaks based on disease surveillance, meteorological and socio-economic data. BMC Infect. Dis. 2019;19:272. doi: 10.1186/s12879-019-3874-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saswat T, Kumar A, Kumar S, Mamidi P, Muduli S, Debata NK, Pal NS, Pratheek BM, Chattopadhyay S, Chattopadhyay S. High rates of co-infection of Dengue and Chikungunya virus in Odisha and Maharashtra, India during 2013. Infect. Genet. Evol. 2015;35:134–141. doi: 10.1016/j.meegid.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 13.Grmanová G, Laurinec P, Rozinajová V, Ezzeddine AB, Lucká M, Lacko P, Vrablecová P, Návrat P. Incremental ensemble learning for electricity load forecasting. Acta Polytech. Hung. 2016;13(2):97–117. [Google Scholar]
- 14.Rios RA, Rios TN, Melo R, de Santana ES, Carneiro TMS, Junior ADO. Applying concept drift to understand hepatitis evolution in Brazil. Cybern. Syst. 2020;51(6):631–645. doi: 10.1080/01969722.2020.1724640. [DOI] [Google Scholar]
- 15.Ejigu BA, Wencheko E, Moraga P, Giorgi E. Geostatistical methods for modelling non-stationary patterns in disease risk. Spat. Stat. 2020;35:100397. doi: 10.1016/j.spasta.2019.100397. [DOI] [Google Scholar]
- 16.Sultana N, Sharma N, Sharma KP, Verma S. A sequential ensemble model for communicable disease forecasting. Curr. Bioinform. 2020;15(4):309–317. doi: 10.2174/1574893614666191202153824. [DOI] [Google Scholar]
- 17.Papageorgiou EI, Kannappan A. Fuzzy cognitive map ensemble learning paradigm to solve classification problems: application to autism identification. Appl. Soft Comput. J. 2012;12:3798–3809. doi: 10.1016/j.asoc.2012.03.064. [DOI] [Google Scholar]
- 18.Smith ME, Singh BK, Irvine MA, Stolk WA, Subramanian S, Hollingsworth TD, et al. Predicting lymphatic filariasis transmission and elimination dynamics using a multi-model ensemble framework. Epidemics. 2017;18:16–28. doi: 10.1016/j.epidem.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin H, Jha NK. A health decision support system for disease diagnosis based on wearable medical sensors and machine learning ensembles. IEEE Trans. Multi Scale Comput. Syst. 2017;3(4):228–241. doi: 10.1109/TMSCS.2017.2710194. [DOI] [Google Scholar]
- 20.Viboud C, Sun K, Gaffey R, Ajelli M, Fumanelli L, Merler S, Zhang Q, Chowell G, Simonsen L, Vespignani A. The RAPIDD ebola forecasting challenge: synthesis and lessons learnt. Epidemics. 2018;22:13–21. doi: 10.1016/j.epidem.2017.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaman J, Karspeck A. Forecasting seasonal outbreaks of influenza. Proc. Natl. Acad. Sci. USA. 2012;109:20425–20430. doi: 10.1073/pnas.1208772109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarebski AE, Dawson P, McCaw JM, Moss R. Model selection for seasonal influenza forecasting. Infect. Dis. Model. 2017;2:56–70. doi: 10.1016/j.idm.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cobb L, Krishnamurthy A, Mandel J, Beezley JD. Bayesian tracking of emerging epidemics using ensemble optimal statistical interpolation. Spat. Spatiotemporal Epidemiol. 2014;10:39–48. doi: 10.1016/j.sste.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Narula P, Piratla V, Bansal A, Azad S, Lio P. Parameter estimation of tuberculosis transmission model using ensemble Kalman filter across Indian states and union territories. Infect. Dis. Health. 2016;21:184–191. doi: 10.1016/j.idh.2016.11.001. [DOI] [Google Scholar]
- 25.Yang W, Karspeck A, Shaman J. Comparison of filtering methods for the modeling and retrospective forecasting of influenza epidemics. PLoS Comput. Biol. 2014;10:e1003583. doi: 10.1371/journal.pcbi.1003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin S, Su L, Ullah A. Robustify financial time series forecasting with bagging. Econ. Rev. 2014;33(5–6):575–605. doi: 10.1080/07474938.2013.825142. [DOI] [Google Scholar]
- 27.Fang X, Liu W, Ai J, et al. Forecasting incidence of infectious diarrhea using random forest in Jiangsu Province, China. BMC Infect. Dis. 2020;20:222. doi: 10.1186/s12879-020-4930-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rathore SS, Kumar S. Towards an ensemble based system for predicting the number of software faults. Expert Syst. Appl. 2017;82:357–382. doi: 10.1016/j.eswa.2017.04.014. [DOI] [Google Scholar]
- 29.Rathore SS, Kumar S. Linear and non-linear heterogeneous ensemble methods to predict the number of faults in software systems. Knowl. Based Syst. 2017;119:232–256. doi: 10.1016/j.knosys.2016.12.017. [DOI] [Google Scholar]
- 30.Rathore SS, Kumar S. An approach for the prediction of number of software faults based on the dynamic selection of learning techniques. IEEE Trans. Reliab. 2018;68(1):216–236. doi: 10.1109/TR.2018.2864206. [DOI] [Google Scholar]
- 31.Saadallah, A., Priebe, F., Morik, K.: A drift-based dynamic ensemble members selection using clustering for time series forecasting. In: Joint European Conference on Machine Learning and Knowledge Discovery in Databases. pp. 678–694. Springer, Cham (2019)
- 32.Cerqueira, V., Torgo, L., Pinto, F., Soares, C.: Arbitrated ensemble for time series forecasting. In: Joint European Conference on Machine Learning and Knowledge Discovery in Databases. pp. 478–494. Springer, Cham (2017)
- 33.Sharma, N., Juneja, A.: Combining of random forest estimates using LSboost for stock market index prediction. In: 2017 2nd International Conference for Convergence in Technology (I2CT), pp. 1199–1202. IEEE (2017)