Figure 3.
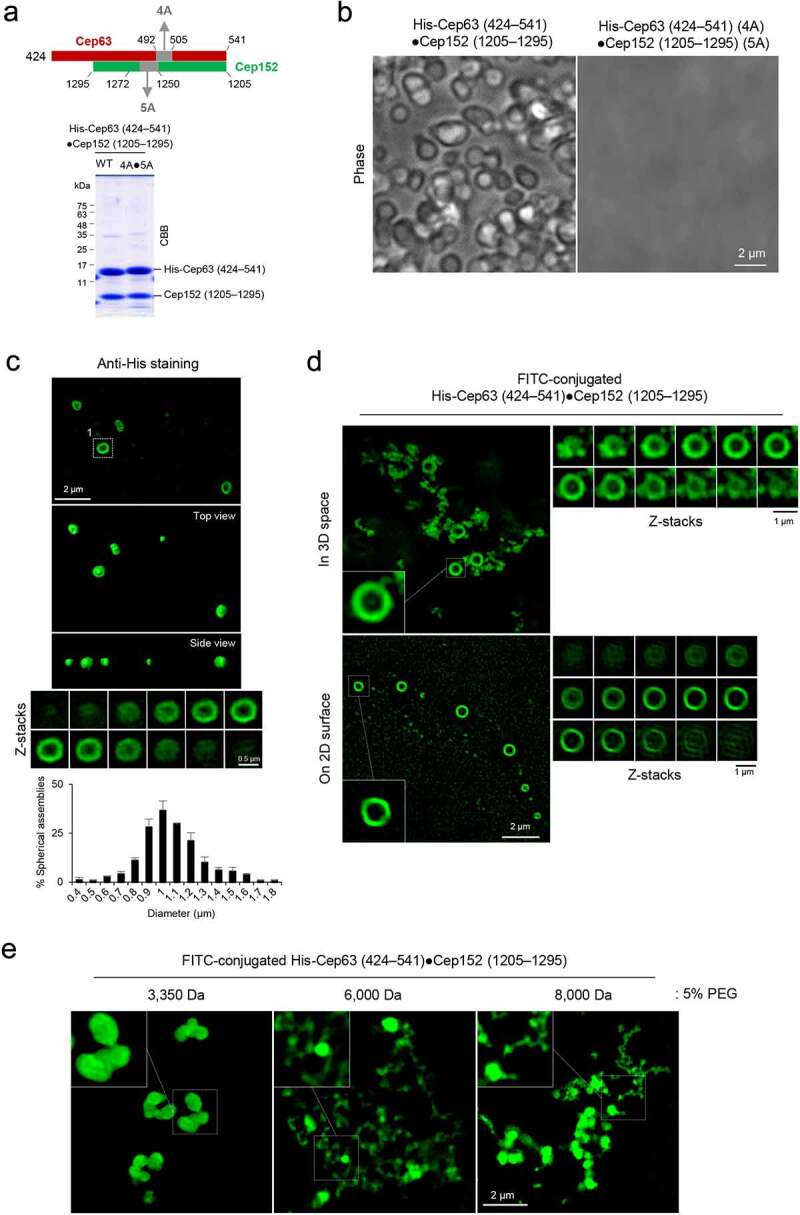
Formation of two morphologically distinct Cep63•Cep152 self-assemblies in vitro. (a), Schematic diagram showing the Cep63 (424–541)•Cep152 (1205–1295) complex and its corresponding 4A•5A hydrophobic mutant and purified proteins stained with Coomassie Brilliant Blue (CBB). (b and c), The proteins (125 μM) in (a) were incubated in an assembly buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5 mM TCEP) + 5% glycerol at 4 °C overnight and subjected to bright-field microscopy (b). The self-assemblies were stained with an anti-His antibody to confirm that spherical condensates are made of the His-Cep63 (424–541)•Cep152 (1205–1295) complex (c). A series of z-stack images in (c) revealing a spherical morphology is shown. The diameters of spherical assemblies were quantified and the results are shown in mean ± s.d. (n = 321 condensates obtained from two independent experiments). (d), 3D-SIM analysis of FITC-conjugated His-Cep63 (424–541)•Cep152 (1205–1295) self-assemblies generated either in a PCR tube (i.e., 3D space) or on a poly-L-lysine-coated slide glass (i.e., 2D surface) for 14 hours. Right panels, a series of z-stack images showing either a spherical (top) or cylindrical (bottom) morphology. Note of the different diameters for different z-stacks of the spherical assembly (top). 3D surface-rendered movies are provided in Movie S3. (e), 3D-SIM analysis of His-Cep63 (424–541)•Cep152 (1205–1295) self-assemblies generated in a 3D space in the presence of the indicated 5% polyethylene glycols (PEGs) for 14 hours
