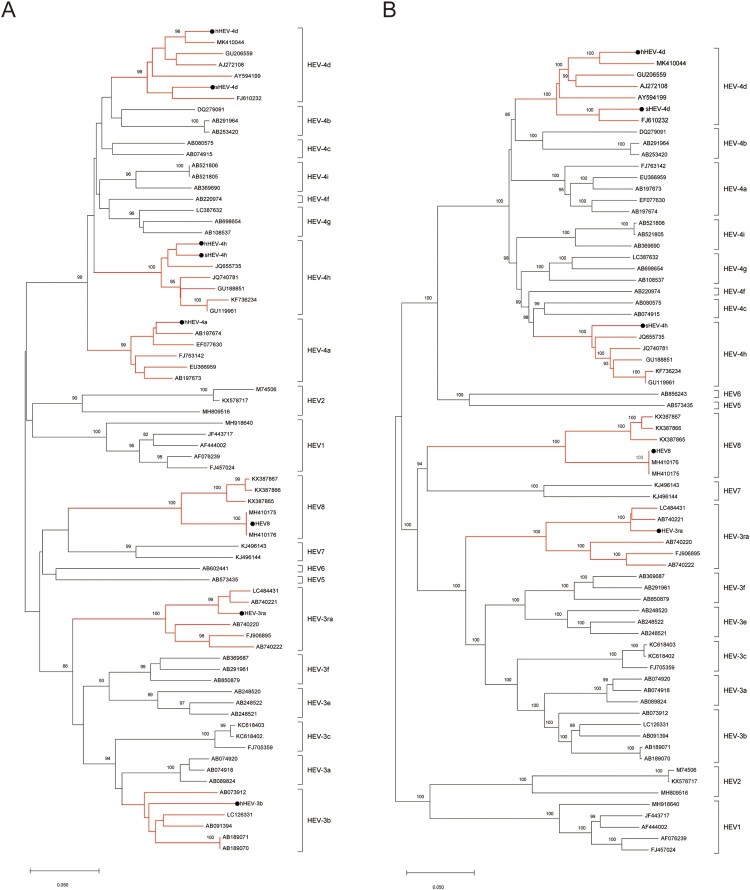
Figure 1.
Phylogenetic analysis of the strains used to infect rabbits. (A) Phylogenetic analysis of the eight strains based on a partial nucleotide sequence about 350 bp of the ORF2 region. (B) Phylogenetic analysis of nearly complete sequences of five strains (HEV-3ra, hHEV-4d, sHEV-4d, sHEV-4h and HEV8). The complete or nearly complete sequences of the above five virus strains used in the study were obtained. Sixty-six ICTV proposed reference strains were used and the GenBank accession numbers of all reference sequences are shown in the figure. The phylogenetic trees were constructed by the neighbour-joining method. Numbers indicate nodes where bootstrap support was >80% out of 1000 replicates. Bar: nucleotide sequence distances; Dots: strains used to infect rabbits in the study. GenBank numbers: hHEV-3b, MF996356; hHEV-4a, KP325707; hHEV-4d, MT993748; hHEV-4h, KP325697; sHEV-4d, MT993749; sHEV-4h, MT993750; HEV-3ra, JX109834; HEV8, MH410174.