Fig. 4.
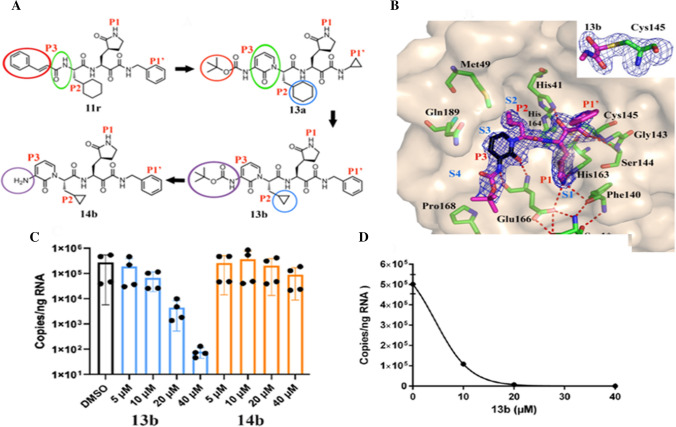
a Chemical structures of α-ketoamide inhibitors 11r, 13a, 13b and 14b. Coloured circles feature the modifications from one development step to the next (see detail in text); b compound 13b in the substrate-binding cleft positioned between domains I and II of the Mpro. The Fo-Fc density is manifested for the inhibitor. Carbon atoms of the inhibitor are magenta coloured, but black in the pyridone ring; nitrogen atoms are blue, oxygens are red and sulphur yellow. S1, S2, S3 and S4 subunits (light blue) indicate the canonical binding pockets for P1, P2, P3 and P4 units (red) of the peptidomimetic inhibitor. Dashed red lines indicate hydrogen bonds. The interaction between the N-terminal residue of chain B, Ser1* and Glu166 of chain A is necessary for preserving the S1 pocket in the precise shape and the enzyme in the functional form. Inset represents the formation of thiohemiacetal by the nucleophilic attack of the catalytic cysteine onto the α-carbon of the inhibitor; c and d inhibition of SARS-CoV-2 replication by 13b inhibitor in human Calu3 lung cells; c SARS-CoV-2-infected Calu-3 cells stimulated with DMSO (black bar) and varied amounts (5, 10, 20 and 40 μM) of 13b (blue bars), and 14b (orange bars) evaluated at 24 h; d a dose–response curve for the estimation of the EC50 value of the inhibitor 13b against SARS-CoV-2. Reprinted from Ref. [75]. Copyright 2020 American Association for the Advancement of Science