Abstract
The complete mitochondrial and plastid genomes of the microalga Pavlova lutheri strain NIVA-4/92 are reported. The circular-mapping mitogenome is 36,202 bp in length, contains 22 protein-coding genes, 24 tRNAs, and has a GC content of 37.5%. Like other haptophytes the mitogenome contains a single large, complex repeat region of approximately 5.4 kbp. The plastome is 95,281 bp in length and has a GC content of 35.6%. It contains 111 protein-coding genes and 27 tRNAs.
Keywords: Haptophyte, metabolic model, aquaculture, lipid metabolism, DHA
The microalga Pavlova (Pavlovophyceae) is a rich source of long-chain polyunsaturated fatty acids. Long-used in the aquaculture industry as a live feed, Pavlova synthesizes high proportions of docosahexaenoic acid (DHA), eicosapentaenoic acid (EPA), and a set of unique sterols. The Pavlovophyceae comprises a group of four genera and at least 13 characterized species that typically branch some genetic distance from other haptophytes (Liu et al. 2009; Bendif et al. 2011). They have a red-alga derived plastid acquired via secondary endosymbiosis, and despite their biogeochemical and industrial significance are under-represented in genomic studies (Baurain et al. 2010). Here we report the complete mitogenome and plastome of Pavlova sp. NIVA-4/92, which is available from the Norwegian Culture Collection of Algae (NORCCA) and reportedly originates from Oslofjord, Norway (59°21′N,10°33′E).
High molecular weight DNA was sequenced on a Pacific Biosciences Sequel system by Arizona Genomics Institute (Tucson, Arizona USA). We assembled the whole genome with Canu version 1.7 (Koren et al. 2017), including complete circular-mapping mitochondrial and plastid genome contigs. The sequences were polished to high accuracy with Blasr and Arrow command-line tools from SMRT Link version 5.1 (Pacific Biosciences, Menlo Park, California USA). To ensure there were no remaining indels, 250 bp paired-end Illumina reads were aligned to the genomes with BWA-MEM and the sequences were verified with Pilon (Walker et al. 2014) and FreeBayes (Garrison and Marth 2012). Sequence annotation was assisted by a partial Pavlova lutheri mitogenome sequence (HQ908424.1) in addition to GeSeq (Tillich et al. 2017), tRNAscan-SE version 2.0.3 (Chan and Lowe 2019), RNAweasel (http://megasun.bch.umontreal.ca/RNAweasel) and assembled RNA-seq transcripts.
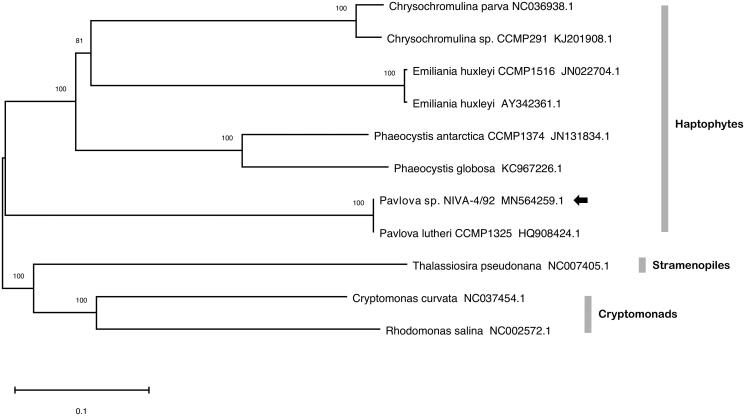
The mitochondrial genome (MN564259.1) is 36,202 bp in length, has a GC content of 37.46%, encodes 22 protein-coding sequences, and 24 tRNAs. It contains a single 5.4 kbp repeat region, a feature found in other haptophyte mitogenomes including Emiliania huxleyi (2 kbp repeat region) and Chrysochromulina sp. CCMP291 (9.5 kbp repeat region). Analysis with EMBOSS einverted (Rice et al. 2000) indicates that the repetitive region contains a pair of inverted sequences 1846 and 2042 bp in length that share 85.7% identity. Tandem repeats finder (Benson 1999) identified 41 repeat sequences that extend through 5295 bp of the same region. As shown in Figure 1, the genus Pavlova forms the outermost branch amongst haptophytes and the mitogenome coding sequences of NIVA-4/92 are identical to those of Pavlova lutheri CCMP1325. The plastome of NIVA-4/92 (MT364382.1) is 95,281 bp, has a GC content of 35.60%, contains 111 protein-coding sequences, and 27 tRNAs. Its identity with CCMP1325 confirms that NIVA-4/92 is Pavlova lutheri. In connection with the nuclear genome, the mitogenome and plastome sequences will facilitate analysis of organelle bioenergetics, transcription, signaling, construction of compartmentalized genome-scale metabolic models, and potentially aid chloroplast transformation in this industrially significant microalga.
Figure 1.
Phylogenetic analysis of 11 mitochondrial genomes with 15 protein-coding genes common to all strains, including eight haptophytes, two cryptomonads, and a diatom. Sequences were aligned with Clustal Omega (Sievers et al. 2011), prepared with GBlocks (Talavera and Castresana 2007), and concatenated to a length of 11,594 nucleotide positions. Tree construction was performed in MEGA-X with neighbor-joining and 1000 bootstrap replications. Units are substitutions per site and support values are indicated.
Funding Statement
CJH is supported by the European Union’s Horizon 2020 research and innovation program under Marie Skłodowska-Curie grant agreement number [749910]. CJH thanks the National Center for Genome Resources (NCGR) New Mexico, USA, Colorado School of Mines (Golden, CO, USA), and Sigma2 Uninett (Norwegian national computing infrastructure) project number [NN9634K] for computing support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference numbers MN564259.1 and MT364382.1.
References
- Baurain D, Brinkmann H, Petersen J, Rodríguez-Ezpeleta N, Stechmann A, Demoulin V, Roger AJ, Burger G, Lang BF, Philippe H.. 2010. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Biol Evol. 27(7):1698–1709. [DOI] [PubMed] [Google Scholar]
- Bendif EM, Probert I, Hervé A, Billard C, Goux D, Lelong C, Cadoret J-P, Véron B.. 2011. Integrative taxonomy of the Pavlovophyceae (Haptophyta): a reassessment. Protist. 162(5):738–761. [DOI] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27(2):573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Chan PP, Lowe TM.. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Gene prediction. New York: Humana; p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G.. 2012. Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907 [q-bio.GN] [Google Scholar]
- Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM.. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27(5):722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Probert I, Uitz J, Claustre H, Aris-Brosou S, Frada M, Not F, de Vargas C.. 2009. Extreme diversity in noncalcifying haptophytes explains a major pigment paradox in open oceans. Proc Natl Acad Sci USA. 106(31):12803–12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European molecular biology open software suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7(1):539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S.. 2017. GeSeq- versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. . 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/genbank/, reference numbers MN564259.1 and MT364382.1.