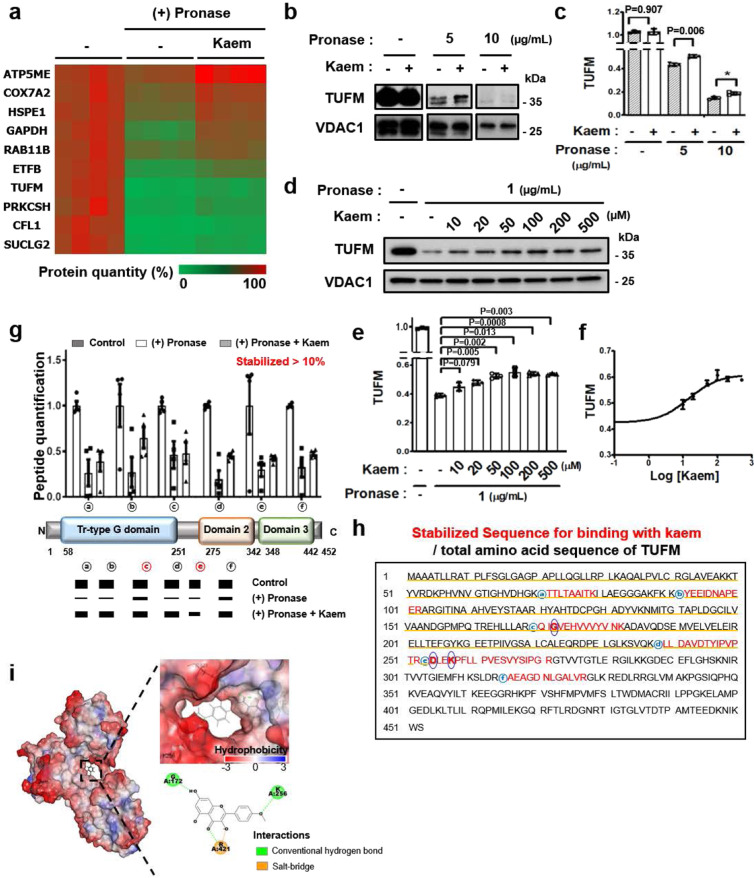
Fig. 5. DARTS LC–MS quantitative proteomic analysis indicates TUFM is a biophysical target of Kaem.
a Heatmap of top 10 proteins showing significantly increased stability to proteolysis upon Kaem treatment. 3T3-L1 cell lysate was treated with pronase for 10 min with/without Kaem pretreatment. SWATH analysis was conducted to identify proteins with varying protease stability (by KBSI). Top 10 protein target candidates were selected according to the sequential criteria based on the amount of detected peptide. b, c DARTS assay for target validation. 3T3-L1 cell lysate was treated with pronase for 10 min with/without Kaem pretreatment and subjected to western blot analysis using anti-TUFM and anti-VDAC1 antibodies. Representative images (b) and intensity of TUFM immunoblot bands (c). The blots were processed in parallel. Kaem, 2 mM. Graph shows mean ± SD from three independent experiments. d–f 3T3-L1 cells were treated with pronase for 10 min with/without Kaem pretreatment in a dose-dependent manner, then subjected to western blot analysis using anti-TUFM and anti-VDAC1 antibodies. Representative images (d), the intensity of TUFM immunoblot bands (e), and sigmoidal curve (f). Graph shows mean ± SD from three independent experiments. g Graph of top isotope quantitation (TIQ) for TUFM peptides (upper). Schematic illustration of the peptide-locus of TUFM consisting of the tr-type G domain, domain 2, and domain 3 (lower). Graph shows mean ± SEM of each peptide quantification (n = 4). h Complete amino acid sequence of TUFM. Core amino acids correlated with in silico docking analysis are indicated by blue circles. i In silico docking model of Kaem directly interacting with TUFM (PDB ID: 1XB2, interaction energy: 44.9 kcal/mol). Presumably binding amino acids and interaction mode are depicted. Statistical significance was assessed by the Student’s t-test. ***P < 0.001; **P < 0.01; *P < 0.05.