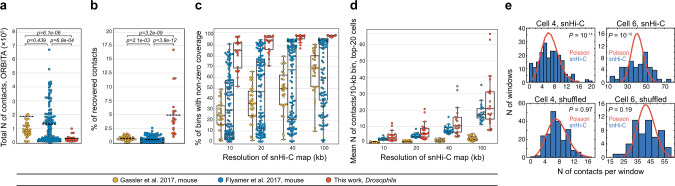
Fig. 2. snHi-C datasets in Drosophila represent a major portion of the genome and are not random matrices.
a Number of ORBITA-captured contacts per individual nuclei obtained for Drosophila in the current work, compared with mouse oocytes from Flyamer et al.32 and G2 zygotes pronuclei from Gassler et al.34. **p < 0.01 using the Mann–Whitney two-sided test. n = 20, 120, and 32 nuclei for Drosophila in the current work, mouse oocytes from Flyamer et al.32 and G2 zygotes pronuclei from Gassler et al.34, respectively (the same is true for (b) and (c)). b Percentage of recovered contacts out of the total possible for Drosophila in the current work, compared with mouse oocytes from Flyamer et al.32 and G2 zygotes pronuclei from Gassler et al.34. P-values are calculated using the Mann–Whitney two-sided test. c Percentage of bins with non-zero coverage for autosomes and sex chromosome of Drosophila, murine oocytes, and G2 zygote pronuclei. Boxplots represent the median, interquartile range, maximum and minimum. d Mean number of contacts per 10-kb genomic bin in top-20 cells in the current work, compared with mouse oocytes from Flyamer et al.32 and G2 zygotes pronuclei from Gassler et al.34. Boxplots represent the median, interquartile range, maximum and minimum. e Distributions of the number of contacts in windows of fixed size (100 kb for the Cell 4, and 400 kb for the Cell 6; chr2R) in snHi-C data and shuffled maps for two individual cells (blue bars). The red curve shows the Poisson distribution expected for an entirely random matrix with the same number of contacts. P-values were estimated by the goodness of fit test. n = 211 and 52 windows for the cell 4 and for the cell 6, respectively.