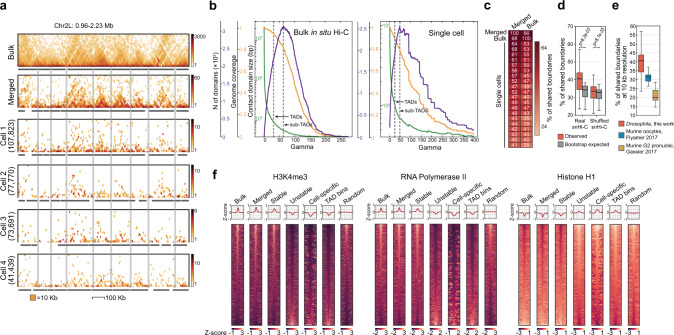
Fig. 3. Stable TAD boundaries are defined by high level of active epigenetic marks.
a Example of a genomic region on Chromosome 2L with a high similarity of TAD profiles (black rectangles) in individual cells and bulk BG3 in situ Hi-C data. Number of unique captured contacts is shown in brackets. Positions of TAD boundaries identified in bulk BG3 in situ Hi-C data (top panel) are highlighted with gray lines. Here and below, TADs are identified using lavaburst software. b Dependence of the contact domain (CD) size (green), genome coverage by CDs (orange), and number of identified CDs (violet) on the γ value in bulk (left) and single-cell (right) BG3 Hi-C data. γ values selected for the calling of sub-TADs (γmax) and TADs (γmax/2) are marked with vertical gray lines. c Percentage of TAD boundaries shared between single cells, bulk BG3 in situ Hi-C, and merged snHi-C data. d Percentage of shared boundaries in real snHi-C, shuffled control maps, and bootstrap expected. Boxplots represent the median, interquartile range, maximum and minimum. **p < 0.01 using the Mann–Whitney two-sided test. n = 380 comparisons between individual cells. e Percentage of shared boundaries in real snHi-C for Drosophila, murine oocytes from Flyamer et al.32 and G2 zygote pronuclei from Gassler et al.34. Boxplots represent the median, interquartile range, maximum and minimum. n = 380 comparisons between individual cells. f Heatmaps of active (H3K4me3, RNA Polymerase II) and inactive (H1 histone) chromatin marks centered at single-cell TAD boundaries from different groups (±100 kb). Bulk—conventional BG3 in situ Hi-C; merged—aggregated snHi-C data from all individual cells; stable and unstable—boundaries found in more and in less than 50% of cells, respectively; cell-specific—boundaries identified in any one individual cell; TAD bins—genomic bins from TAD interior; random—randomly selected genomic bins.