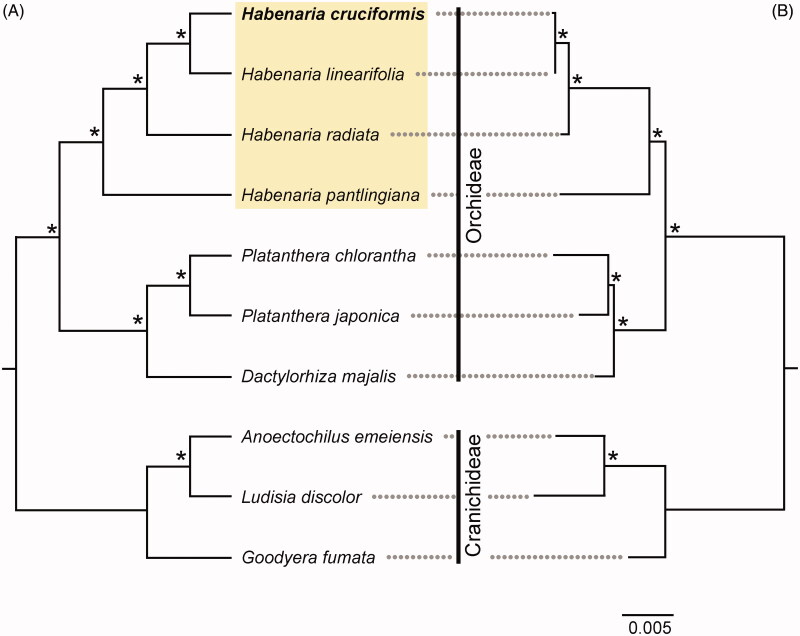
Figure 1.
Phylogenetic trees resulting from (A) maximum parsimony (MP) and (B) Bayesian inference (BI) analyses of chloroplast 76 protein coding genes. Four Habenaria species are highlighted in yellow. Asterisks near nodes indicate bootstrap support = 100% for MP analysis and posterior probability = 1.00 for BI analysis. The bar represents 0.005 nucleotide substitution per site. GenBank accession numbers: Anoectochilus emeiensis: NC033895; Dactylorhiza majalis: NC044644; Goodyera fumata: NC026773; Habenaria cruciformis: MT863537; Habenaria linearfolia: MT863538; Habenaria pantilingiana: NC026775; Habenaria radiata: NC035834; Platanthera chlorantha: NC044626; Platanthera japonica: NC037440; Ludisia discolor: NC030540.