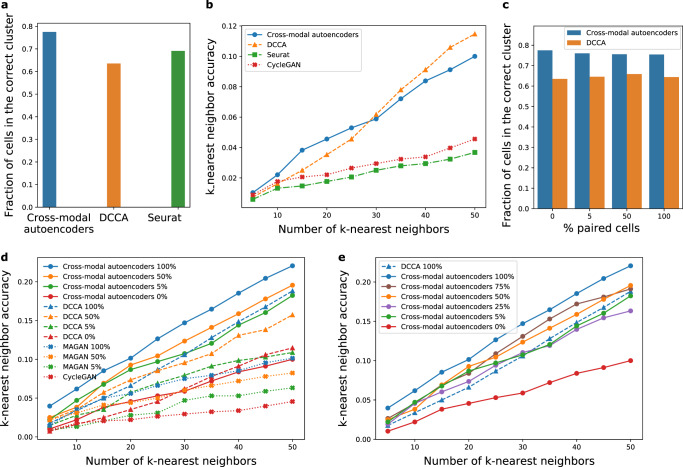
Fig. 2. Performance of our multimodal data integration method (cross-modal autoencoders), deep canonical correlation analysis (DCCA), Seurat, CycleGAN, and MAGAN on paired RNA-seq and ATAC-seq data.
a Fraction of cells that were assigned to the correct treatment time cluster based on their embedding in the integrated latent space that was learned by fitting our cross-modal autoencoder model, DCCA, or Seurat. b k-nearest neighbor accuracy for quantifying the quality of matching between local neighborhoods for our cross-modal autoencoder model, DCCA, Seurat, and CycleGAN trained with 0% supervision (no paired samples). c Fraction of cells that were assigned to the correct treatment time cluster for our cross-modal autoencoders and DCCA trained with varying amount of paired samples. d k-nearest neighbor accuracy for our cross-modal autoencoders, DCCA, MAGAN, and CycleGAN trained with 0, 5, 50, and 100% of the paired samples. e k-nearest neighbor accuracy for our cross-modal autoencoder model trained with varying amount of paired samples versus DCCA trained on all paired samples. In a–c colors denote different domain translation methods and in d–e colors denote different levels of supervision (paired samples). Additionally, different markers denote different domain translation methods.