Abstract
Human herpesvirus 6A and 6B (HHV-6) can integrate into the germline, and as a result, ∼70 million people harbor the genome of one of these viruses in every cell of their body. Until now, it has been largely unknown if 1) these integrations are ancient, 2) if they still occur, and 3) whether circulating virus strains differ from integrated ones. Here, we used next-generation sequencing and mining of public human genome data sets to generate the largest and most diverse collection of circulating and integrated HHV-6 genomes studied to date. In genomes of geographically dispersed, only distantly related people, we identified clades of integrated viruses that originated from a single ancestral event, confirming this with fluorescent in situ hybridization to directly observe the integration locus. In contrast to HHV-6B, circulating and integrated HHV-6A sequences form distinct clades, arguing against ongoing integration of circulating HHV-6A or “reactivation” of integrated HHV-6A. Taken together, our study provides the first comprehensive picture of the evolution of HHV-6, and reveals that integration of heritable HHV-6 has occurred since the time of, if not before, human migrations out of Africa.
Keywords: human herpesvirus 6, phylogenetics, genomics, paleovirology, telomere biology
Introduction
Viral sequences can become integrated into the host genome, either as part of their replication strategy or through host-mediated recombination. When this occurs in germline cells, individuals can arise harboring the virus in every cell of their body, and transmit it to their offspring in a Mendelian fashion (Katzourakis and Gifford 2010; Aswad and Katzourakis 2016). These endogenous viral elements (EVEs) can eventually reach fixation in the population and persist for millions of years (Katzourakis et al. 2009).
Such ancient integrations are an invaluable resource for studying the long-term evolution of viruses and the evolutionary dynamics with their hosts. However, far less is known about the early stages of endogenization—before an EVE has reached fixation—because this requires large-scale genomic screening at the population level. For this reason, only a handful of unfixed EVEs have been identified, such as the cancer-inducing koala retrovirus (Tarlinton et al. 2006), or the HERVK(HML2) group of endogenous retroviruses, which are insertionally polymorphic in humans in different populations (Wildschutte et al. 2016).
Among the most notable unfixed EVEs are the roseoloviruses human herpesvirus 6A and 6B (HHV-6), which are found in ∼1% of the human population (Pellett et al. 2012). In contrast to these EVEs, the closely related circulating strains of HHV-6A and 6B are extremely widespread. For instance, the seroprevalence of HHV-6B is over 90% worldwide (Kaufer and Flamand 2014; Kühl et al. 2015). Primary infection occurs in infants under the age of three, and typically presents with a high fever and rash, and complications include febrile seizures and encephalitis (Hall et al. 1994; Mohammadpour Touserkani et al. 2017).
Like most herpesviruses, HHV-6A and 6B establish life-long latency but can reactivate resulting in virus replication. HHV-6 reactivation has been implicated in a number of diseases including encephalitis and graft rejection in transplant patients (Pantry et al. 2013; Hill et al. 2016). HHV-6A and 6B integrate their genomes into the telomeres of latently infected cells, possibly as a strategy to maintain their genomes during latency. This feature of their viral replication cycle, unique among HHVs, could explain why they are the only endogenous HHVs identified (a similar Roseolovirus has been identified in the genome of the Philippine Tarsier) (Aswad and Katzourakis 2014). In its endogenous form, the virus is described in the literature as inherited chromosomally integrated HHV-6 (iciHHV-6). Previous reports on iciHHV-6 have used pedigrees to demonstrate inheritance of iciHHV-6 (Huang et al. 2014), but the deeper evolutionary history of iciHHV-6 has thus far only been performed on relatively small data sets (Zhang et al. 2017).
Reactivation of HHV-6 and iciHHV-6 has been confirmed by a number of studies experimentally as well as from evidence in iciHHV-6 positive patients (Hall et al. 2010; Gravel, Hall, et al. 2013; Prusty et al. 2013; Huang et al. 2014; Kühl et al. 2015). Recent work has demonstrated a link between likely iciHHV-6 reactivation in patients with various cardiovascular and myocardial diseases, including angina pectoris, chronic heart failure in adults, and a case of neonatal dilated cardiomyopathy (Das 2015; Gravel et al. 2015; Kühl et al. 2015). Multiple other associations between HHV-6/iciHHV-6 and disease have been documented ranging from graft rejection in transplant patients to Alzheimer’s disease, but the causal role of HHV-6A or 6B remains uncertain (Hill et al. 2017).
In order to understand the relationship of iciHHV-6 to the onset and/or progression of specific diseases, there are a number of crucial questions that need to be tackled first. For instance, we do not know if and how iciHHV6 differs from circulating viral strains, or if there is a difference between the integration mechanism for HHV-6A and HHV-6B. Moreover, we do not know whether germline integrations are still occurring, or whether the 1% of iciHHV6 carriers represents a limited number of ancient events that expanded to their current prevalence.
There has been an increasing number of iciHHV6 genome sequences available, thanks to the development of enrichment techniques that use an Illumina-based approach (Depledge et al. 2011; Brown et al. 2016; Greninger, Knudsen, et al. 2018). However, these data do not allow the identification of the chromosomal location of the virus due to the short length of NGS reads and the fact that the virus integrates into difficult-to-sequence host telomeres.
One direct approach to identifying the chromosomal location of the iciHHV-6 genome is by fluorescence in situ hybridization (FISH), but this approach is laborious, expensive, and requires considerable technical expertise. Thus far, iciHHV-6 has been identified in several chromosomes, with certain loci recurring more often than others (e.g., 17p and 22q) (Osterrieder et al. 2014). It is important to understand whether a bias for certain chromosomes exists in order to investigate the reasons and effects of such a bias, which may be linked to disease phenotypes.
Given the nature of the challenges associated with studying iciHHV-6, we set out to develop a phylogenetic framework to address these basic questions about the evolution and natural history of this phenomenon. In addition to collating existing HHV-6 sequencing data, we sequenced additional patients and mined public human genomes to identify novel integrations. Our conclusions are strengthened by corroborating FISH experiments that specify the chromosomal integration loci of the major global clades of endogenous HHV-6 which we identify here.
Results
We collected sequences from 11 papers published between 1999 and 2018 containing 84 circulating HHV-6 and 112 iciHHV-6 genome sequence (Gompels et al. 1995; Dominguez et al. 1999; Isegawa et al. 1999; Gravel, Ablashi, et al. 2013; Tweedy et al. 2015, 2016; Zhang et al. 2016, 2017; Greninger, Knudsen, et al. 2018; Greninger, Roychoudhury, Makhsous, et al. 2018; Telford et al. 2018) (table 1). To expand this data set, we performed targeted NGS HHV-6 sequencing on subjects previously identified to carry iciHHV-6: 33 samples from a chronic heart failure cohort and 25 samples from a study of preeclampsia study. We developed a new bioinformatic mining technique for NCBI sequence read archive (SRA) that allowed identification of 97 records with HHV-6 read depth suggestive of iciHHV-6. Seven of these could be assembled into near full-length HHV6A/B genomes. Sample information for all sequences used in this study can be found in supplementary table S1, Supplementary Material online.
Table 1.
Sources for Existing Sequences That Were Reanalyzed as Part of the Data Set for This Study.
| Date | HHV-6A | HHV6-B | Strain | Circulating/Endogenous | Publication |
|---|---|---|---|---|---|
| 1999 | 1 | Z29 | Circulating | Dominguez G, et al. J Virol. 73:8040–52 (1999). | |
| 1999 | 1 | HST | Circulating | Isegawa Y, et al. J Virol. 73:8053–8063 (1999). | |
| 1995 | 1 | U1102 | Circulating | Gompels UA, et al. Virology 209:29–51 (1995). | |
| 2013 | 1 | GS | Circulating | Gravel A, Ablashi D, Flamand L. Genome Announc. 1 (2013). | |
| 2015 | 1 | AJ | Circulating | Tweedy J, et al. Genome Announc. 3 (2015). | |
| 2016 | 1 | Endogenous | Tweedy JG, et al. Viruses 8 (2016). | ||
| 2016 | 1 | Endogenous | Zhang E, et al. Sci Rep. 6 (2016). | ||
| 2017 | 6 | 21 | Endogenous | Zhang E, et al. J Virol. 91:JVI.01137-17 (2017). | |
| 2018 | 3 | 6 | Endogenous | Telford M, Navarro A, Santpere G. Sci Rep. 8:3472 (2018). | |
| 2018 | 10 | 125 | 74 endogenous, 60 circulating | Greninger AL, et al. BMC Genomics 19:204 (2018). | |
| 2018 | 9 | 8 | Circulating | Greninger AL, et al. J Virol. 92 (2018). |
Circulating and Integrated HHV6 Have Distinct Evolutionary Histories
To determine if the circulating strains differ from the integrated viruses, we reconstructed the phylogeny of 261 HHV-6 genomes, annotating their source (iciHHV-6 or circulating strain). This generated a strikingly different result for HHV-6A compared with HHV-6B (figs. 1 and 2 andsupplementary figs. S2 and S3, Supplementary Material online). For HHV-6A, the circulating strains (clade A1) and iciHHV-6 sequences belong to distinct clades separated by long internal branches and supported with a posterior probability of 1 (fig. 1). iciHHV-6A genomes at different chromosomal loci and in people from a diverse geographical origin are more closely related to one another than any of them are to the circulating strains (clades A2–4, fig. 1 andsupplementary fig. S2, Supplementary Material online). This phylogenetic pattern indicates that the ancestral circulating strains that resulted in these particular independent integration events are not among the known currently circulating strains sampled here. Similarly, the integrated HHV-6A is not acting as a reservoir for ongoing production of circulating strains. We would stress, however, that future sampling could change either or both of these interpretations.
Fig. 1.
(A) HHV-6A subtree consisting of 13 circulating HHV-6A and 38 iciHHV-6A sequences. Gray numbers at each node represent posterior probabilities, showing only those with >0.80. Green labels represent endogenous iciHHV-6, whereas blue labels represent circulating infectious viruses. Where available, confirmation of the chromosomal location of iciHHV-6 is indicated with red labels. Gray text at each tip describes the geographical source of the sequence as well as the ethnicity of the patient where this information was available. Black labels indicate known reference strains of HHV-6A. Note that the long branch of KT895199.1 means that we cannot be confident about its placement due to evidence of long-branch attraction from the ML tree (supplementary fig. S2, Supplementary Material online). (B) HHV-6 Bayesian phylogenetic tree reconstructed using 261 HHV-6 and iciHHV-6 sequences.
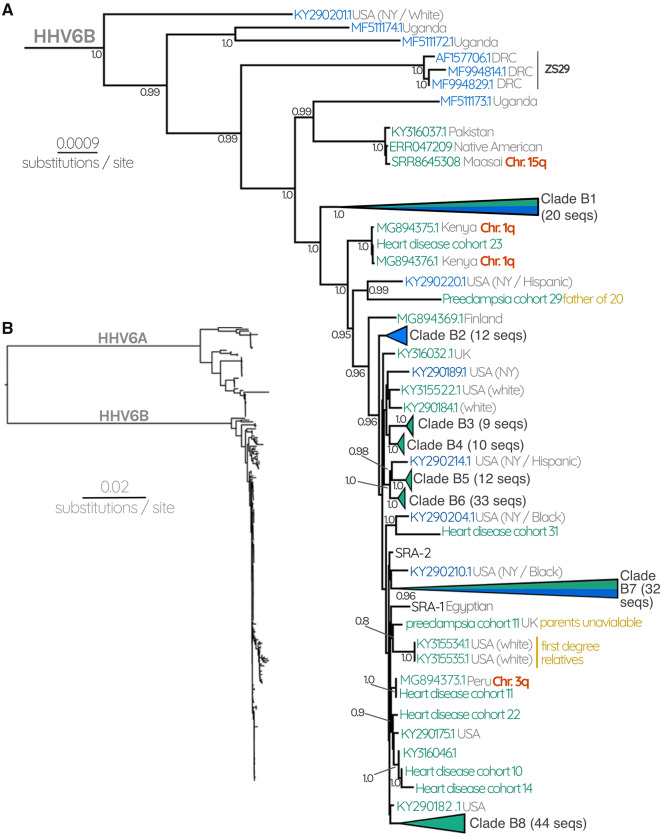
In contrast to HHV-6A, the tree for HHV-6B revealed a more entangled topology between circulating and endogenous genomes, where no phylogenetic segregation between the two was observed (fig. 2 andsupplementary fig. S3, Supplementary Material online). Overall, the branch lengths within the HHV-6B subtree are much shorter than those that separate HHV-6A clades. The iciHHV-6B sequences we observe are within the diversity of the known circulating strains. In spite of the overall lower phylogenetic diversity, we were able to identify specific lineages of circulating HHV-6B strains that are very closely related to the viruses that iciHHV-6B sequences derived from. Specifically, there are two well-supported clades (posterior probability >0.95) composed primarily of circulating strains, but also include endogenous sequences (four in clade B1 and one in B7, figs. 2 and 3). As with HHV-6A, there are “exclusive” clades that only contain either circulating or endogenous viruses, however for HHV-6B these are interspersed throughout the tree, indicating that certain endogenous lineages are more closely related to some circulating lineages than they are to others (fig. 2 andsupplementary fig. S3, Supplementary Material online). This overall topology is also supported by a maximum-likelihood tree constructed using all codon positions of the coding region (supplementary figs. S2 and S3, Supplementary Material online). Taken together, these observations suggest that HHV6B viruses capable of integration are nested within the diversity of currently circulating HHV-6B.
Fig. 2.
(A) HHV-6B subtree consisting of 72 circulating HHV-6B and 137 iciHHV-6B sequences. Gray numbers at each node represent posterior probabilities, showing only those with >0.80. Green labels represent endogenous iciHHV-6, whereas blue labels represent circulating infectious viruses. Collapsed nodes are represented as triangles for clarity (expanded in fig. 3). Collapsed nodes are labeled either green, blue, or both depending on whether the clade consists entirely of iciHHV-6B, HHV-6B, or a mixture of both. Where available, confirmation of the chromosomal location of iciHHV-6 is indicated with red labels. Gray text at each tip describes the geographical source of the sequence as well as the ethnicity of the patient where this information was available. Black labels indicate known reference strains of HHV-6B. (B) HHV-6 Bayesian phylogenetic tree reconstructed using 261 HHV-6 and iciHHV-6 sequences.
In addition to the clades described earlier, the HHV-6B subtree also contains sequences (of both iciHHV-6B and HHV-6B) that are in poorly supported and/or small clades (fig. 2). Some of these may represent integrations that remain at very low prevalence, perhaps because they occurred very recently, or perhaps appear rarely in our data set because they derive from undersampled populations. For instance, a triplet of sequences from two unrelated Kenyan people and one individual of unknown origin grouped with high posterior probability, suggesting an ancestral integration event (MG894375.1, MG894376.1, and heart disease cohort 23, respectively, fig. 2). This is further supported by FISH evidence we generated that demonstrates that both Kenyans harbor the virus in chromosome 1q (fig. 2).
Bioinformatically Identifying iciHHV-6 Chromosomal Locations
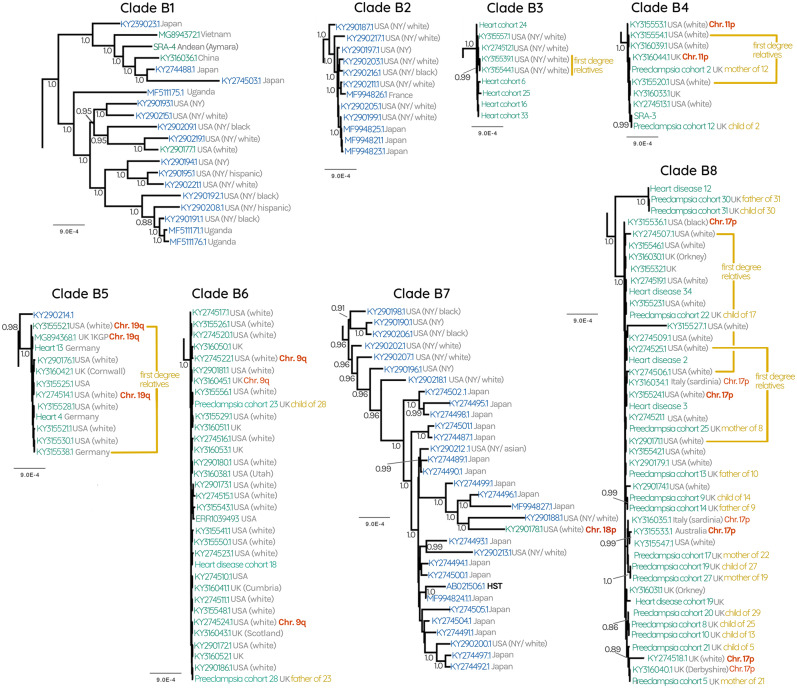
We hypothesized that at least some of the clade structure apparent in the phylogenetic reconstruction is the result of single ancestral events that are stably replicated in the human germline. Such integration events would be identical-by-descent, expanding via human reproduction and linear, vertical transmission, in contrast to expansion by viral replication and horizontal spread. Such endogenous viruses would be predicted to diversify more slowly than expanding during viral replication. The tree evidences such integrations, in the form of monophyletic clades with high posterior probabilities. These clades are characterized by extremely short branch lengths and are separated from one another by relatively longer internal branches, which is particularly clear in the case of HHV-6A clades A2–4 (fig. 1) and HHV-6B clades B3–6 and B8 (figs. 2 and 3). Moreover, the sequences in our data set that derived from families resolve within the same clades, except in the case of the child “Preeclampsia cohort 20,” whose mother was not sequenced and whose father possesses a different iciHHV-6 sequence.
To confirm one prediction of this model, that individuals from these clades indeed have the virus integrated into the same chromosomal location, we performed FISH analyses on 18 cell lines derived from patients whose iciHHV-6 genome is represented in the tree (supplementary fig. S1, Supplementary Material online). In combination with FISH confirmation performed by other groups, we now have direct evidence for the integration locus of 25/177 iciHHV-6 sequences. Across the whole tree, we now know the integration locus for at least one sequence in 11 different clades. We obtained the highest number of confirmations (total seven) for the location of the virus in the HHV-6B clade B8. All of these integrations are located on chromosome 17p, which is therefore almost certainly the site of the integration event that has been expanded via human reproduction to lead to all 41 sequences in clade B8 (fig. 3). The sequences in clades B4, B5, and B6 are almost certainly all represent integrations in chromosomes 11p, 19q, and 9q, respectively. In figure 2, we can infer that the sequences identified by FISH on chromosome 15q, 1q, and 3q are also the locations of the viruses for the other sequences in those smaller clades (fig. 2). For HHV-6A, we infer that clades A2, A3, and A4 are integrations in chromosomes 17p, 19p, and 18q, respectively.
Fig. 3.
HHV-6B subtrees of clades B1–8 collapsed in figure 2. Gray numbers at each node represent posterior probabilities, showing only those with >0.80. Green labels represent endogenous iciHHV-6, blue labels represent circulating infectious viruses. Where available, confirmation of the chromosomal location of iciHHV-6 is indicated with red labels. Gray text at each tip describes the geographical source of the sequence as well as the ethnicity of the patient where this information was available.
Phylogeographic Patterns Reflect Human Migration
Transposable element and EVE insertions can be useful markers to trace human demographic patterns and migrations (Sudmant et al. 2015; Li et al. 2019). Therefore, we next assessed our phylogenetic reconstruction to determine if integrated HHV-6 diversity mirrors the ethnic or geographic distribution of the human hosts. The major clades likely to represent single ancestral integrations are ethnically and/or geographically homogeneous. For instance, among the iciHHV-6A sequences, we observed that individuals from clades A2 and A4 are exclusively European or North American (fig. 1). HHV-6B clades B3–6 and B8 are similarly homogenous and likely represent orthologous integrations in white Europeans and North Americans (and one Australian). This suggests that for each of these clades, those now carrying the virus share a common ancestor who was also European, and thus the virus integration event occurred prior to the diaspora of ancestors of these individuals; the virus thus likely integrated before the colonial era.
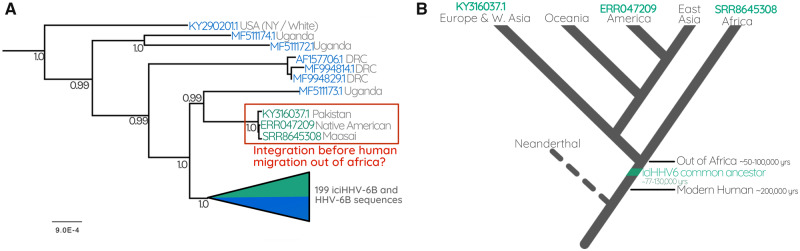
Conversely, our analysis also revealed a previously unidentified Native American carrier of iciHHV-6B, who possesses an HHV-6B sequence distinct from the other North American samples. Instead, this sequence is almost identical to the iciHHV-6B genome of a Maasai Kenyan sequence uncovered through our SRA mining, and a previously identified Pakistani sample (Zhang et al. 2017) (figs. 2 and 4). Unlike the ancestral European integrations, the last common ancestor of these individuals would have been before humans migrated out of Africa (50–100,000 years ago; Nielsen et al. 2017) (fig. 4). The observation that they also resolve near the base of the tree further supports this interpretation, as does the fact that the most closely related sequence outside this clade is a circulating strain isolated from a Ugandan patient (figs. 2 and 4).
Fig. 4.
(A) The Ancient integration of HHV6B. Part of the HHV-6B tree containing the triplet of sequences derived from a Pakistani, Native American, and Maasai Kenyan. (B) A cartoon cladogram indicating the relationships between modern human populations that diverged as humans migrated out of Africa. The model illustrates that the last common ancestor of the three people carrying a near-identical copy of iciHHV-6B must have been before humans left Africa. The cartoon is a simplified interpretation of the model presented in Nielsen et al. (2017).
In addition to the phylogenetic evidence, we performed a dating analysis to examine the age of the different iciHHV-6 clades (fig. 5). Consistent with our interpretation regarding the Maasai–American–Pakistani, we find that the estimated time of this integration to be ∼85,000–342,000 years old (depending on the mutation rate used). Interestingly, the analysis also revealed that iciHHV-6A clades (A2–4) are much older than iciHHV-6B, offering further evidence to support the different evolutionary histories between them. The mean estimates for the integration times of iciHHV-6A genomes are ∼31,000–124,000 years ago, compared with the ∼3,500–14,000 years ago for clades B3–6 and B8.
Fig. 5.
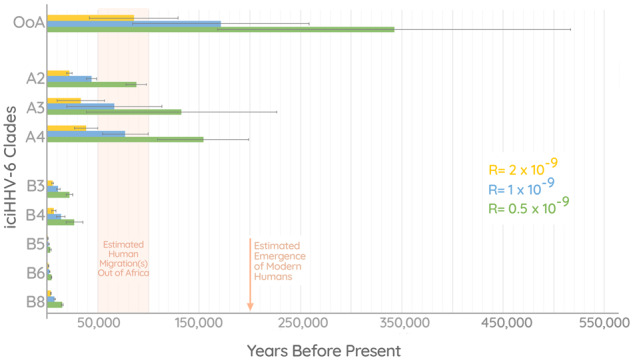
Integration dating analysis. Chart depicting integration date estimates for iciHHV-6 clades A2–4, B3–6, B8, and the “Out of Africa” (OoA) clade consisting of iciHHV-6B sequences from a Maasai Kenyan, Native American, and Pakistani. Each of the colored bars (yellow, blue, and green) represent estimates calculated using different mutation rates as indicated by the key. The error bars shown represent the age estimates based on the upper and lower limits of the 95% confidence interval of the mean pairwise distances in each clade.
The tree also revealed several other clades containing iciHHV-6 sequences from geographically diverse subjects, such as HHV-6B clade B1. This clade includes samples from Uganda, Japan, China, Vietnam, and the Americas (fig. 2). However, a strong case cannot be made for common human ancestry of the iciHHV-6 represented in this clade for two reasons. Firstly, the branch lengths are relatively longer than those of other clades containing iciHHV-6. This indicates a more dynamic evolutionary history than for iciHHV-6 clades for which ancestral integration is the parsimonious interpretation, for example, HHV-6B clades B3–6, B8 and HHV-6A clades A2–4. Secondly, this clade includes both circulating and endogenous viruses. Some of these integrations may be relatively recent events that occurred from strains that remain in circulation. This explanation is particularly compelling in the case of HHV-6B clades B7, where a large group of circulating strains include a single integrated sequence at the tip of the clade, located on chromosome 15p (fig. 3). In contrast to this situation, we do not find examples of rare circulating strains nested within a clade of otherwise endogenous HHV-6, as might be expected if endogenous HHV-6 were acting as a reservoir for ongoing horizontal spread of endemic circulating strains, at least within the limited resolution of the sampling performed to date.
Discussion
We set out to address key questions about the evolution of iciHHV-6 using a phylogenetic approach. We combined this approach with a series of FISH experiments to identify the chromosomal location of the integrated virus for many individuals in the cohort. This analysis revealed several clades that represent single ancestral integration events. Such clades are characterized by extremely short branch lengths that contrast sharply with the between-clade branch length, as well as the longer branch lengths that characterize the evolution of circulating infectious viruses. Moreover, clades of single ancestral integrations include unrelated individuals, from different countries which make it extremely unlikely that they represent multiple integrations of nearly identical strains. This is further corroborated with FISH experiments that consistently support the phylogenetic prediction.
Among the most interesting of these ancestral integrations is one that is represented in our data set by three viral sequences from a Maasai Kenyan, a Pakistani, and Native American. Because the common ancestor of these three people existed before humans emigrated from Africa, we can infer a minimum age for this integration. This interpretation depends on accurate ethnicity information, which we are confident of in this case as all three of these individuals are from well-documented reference panels of projects investigating the population genetics of these groups. The Native American is a reference individual used in a South American study on the influences on physical appearance (Chacón-Duque et al. 2018), the Pakistani sample is from the HGDP-CEPH human diversity reference panel (Bergström et al. 2020), and the Maasai Kenyan is from the international HapMap project (Pemberton et al. 2010). We can therefore be confident that an integration of HHV-6 into anatomically modern humans occurred at least 50–100,000 years ago (Nielsen et al. 2017), which is consistent with the results of the dating analysis, in that all our age estimates are older than 50,000 years.
Calculating the age of the hypothesized OoA integration using the phylogenetically estimated rate of 1× or our arbitrary higher value of 2× offered a plausible time of integration. Using the 0.5× rate based on observed intergenerational SNPs results in an estimate of nearly 350,000 years which would predate the emergence of modern humans. Alternatively, this could be an indication that the true rate of iciHHV-6 is >0.5×.
Identifying the chromosome in which the inherited virus is integrated is a precursor to a wide range of unanswered questions about HHV-6 integration as a human genetic structural variant, such as whether there is a bias for some chromosomes over others or if there are particular phenotypes associated with specific integrations. Although FISH is a relatively reliable method to locate the endogenous virus, it is expensive and time consuming to perform routinely, especially in a clinical setting. Using our rigorous phylogenetic reconstruction allows us to predict the chromosomal location of the virus based on its sequence without performing FISH for every sample, once a certain number of individuals of the clade have been confirmed by FISH. For instance, we can predict with confidence the integration site of individuals bearing clade B8 endogenous HHV-6B (fig. 3), for which terminal chromosome 17p has now been shown to be the cytogenetic locus in seven individuals by FISH. Moreover, there are eight members of this clade derived from parent–offspring pairs, undoubtedly the same integration. Similarly, we have corroborated the chromosome for at least two sequences in clades A2, A3, B4, B5, and B6, as well the small clade consisting of two Kenyan samples and heart disease cohort 23. Therefore, including the inferences, we can make from the tree, we can assign a chromosomal location for 140 different endogenous HHV6 sequences, 79% of all the iciHHV-6 included here.
Our analysis has also revealed the markedly distinct evolutionary history of HHV-6A and HHV-6B. The phylogenetic pattern exhibited in HHV-6A sequences indicates that integrated and circulating viruses diverged long ago, and it may be the case that infectious HHV-6A sequences no longer integrate into the germline or reactivate. This scenario is supported by our dating analysis, which revealed that all three ancestral iciHHV-6A clades are on an average between ∼31,000–124,000 years old (depending on the rate used). In contrast, the average age of iciHHV-6B clades (except for the OoA clade) is ten times lower, between ∼3,500 and 14,000 years (fig. 5). These findings must be confirmed with further sequencing of both circulating HHV-6A and iciHHV-6A, which may reveal a more complex reality that is concealed by undersampling. Nonetheless, integrations of HHV-6B viruses that are within the diversity of the currently circulating strains is a characteristic that sets the virus apart from HHV-6A, even if some HHV-6A strains are eventually shown to still integrate.
On the other hand, the interlaced phylogenetic pattern exhibited in the HHV-6B subtree is consistent with more recent, even ongoing integration in human genomes (such as those in B1 and B7). We attempted to date the examples of such one-off integrations using the divergence of the DR regions that are identical in the exogenous virus (similar to LTR dating for ERVs). However, we found that all the iciHHV-6B sequences that resolved within a clade of exogenous viruses maintained identical DRs. This is supportive of recent integration because the genomes have not had time to accumulate mutations. Furthermore, although iciHHV-6B is capable of reactivation (Prusty et al. 2013; Endo et al. 2014), this is not reflected in the tree topology here. It remains to be seen whether or not reactivation of integrated virus genomes can result in virus transmission, which should become clear with further sampling. This is directly relevant to the safety of blood and organ transplantation from iciHHV-6B donors, where preliminary evidence suggests a higher incidence of graft versus host disease using organs from iciHHV6 donors (Hill and Zerr 2014; Hill et al. 2017).
Current scientific consensus is that HHV-6B exhibits extremely low levels of diversity (Greninger, Knudsen, et al. 2018), and at first glance, our tree seems to confirm this. However, much of this homogeneity can actually be attributed to the large clades of orthologous iciHHV-6 sequences that should be considered as single data points in terms of viral diversity. Moreover, a large proportion of the sequences sampled (both circulating and endogenous) were obtained from white European and North American patients. As viral genomes samples derived from African, Asian, and South American sources exhibit much longer branches, we suspect that the impression of homogeneity will decrease with improved geographical sampling.
Additional sampling is also important to corroborate our phylogenetically inferred conclusions that certain clades represent single ancestral integration events. Given that FISH analyses are not always feasible, developing long-read sequencing approaches that can bridge the virus/subtelomere junction is a priority. This would serve as an alternative source of confirmation that iciHHV-6B genomes sequenced in the future that fall within this clade (or any other) are in fact present at the same chromosomal location. In addition, sampling further diversity will allow us to reconstruct more accurate trees, particularly in the case of HHV-6B. Although we were able to draw important conclusions with this sequence set, much of the deeper structure of the HHV-6B subtree is poorly supported in both the Bayesian reconstruction (fig. 2) and the ML tree (supplementary figs. S2 and S3, Supplementary Material online).
This study developed and applied a conceptually new method to use viral phylogeny to predict the chromosomal location of shared endogenous HHV-6, and showed that this prediction is born out in all cases with available FISH data. Defining endogenous HHV-6 as a discrete set of human genetic structural variants, as this does, is crucial to a range of different questions, such as whether there is a bias toward endogenous HHV-6 integrating or persisting in certain chromosomes (and if so, why), and if different integrations have certain phenotypic effects. The evidence strongly indicates that the integration of HHV-6 has been a feature of human evolution since before our migration out of Africa, which will help us to understand how and why iciHHV-6 sequences have been maintained in the population, and whether such ancient integrations differ in some way from more recent integrations.
Among the most practical of our findings is the observation that HHV-6A viruses are apparently no longer endogenizing (at least in the genomes of Europeans most well represented here) and that iciHHV-6A does not seem to contribute to the pool of circulating HHV-6A. Pending confirmation through further sampling and experimental work, this would be valuable information for medical professionals considering the suitability of blood and organ transplant donors. Conversely, knowing that HHV-6B sequences may still be capable of reactivation will help medical professionals in monitoring posttransplantation patients for potential treatment with antivirals.
Materials and Methods
Sequence Data Collection
To assemble a large data set of HHV-6 genomes, we performed targeted-enrichment Illumina sequencing of 33 heart disease patients confirmed to be carriers of iciHHV6, as well as 25 iciHHV-6 genomes from a preeclampsia study that include mothers, fathers, and their children. iciHHV-6 status was confirmed for samples in this study by identifying a 1:1 ratio of HHV6 genomes to a housekeeping gene through qPCR. In addition, we mined the NCBI SRA to identify seven novel iciHHV-6 sequences in human genome data. To these new sequences, we added 196 publicly available HHV-6 and iciHHV-6 genome sequences that were previously published in 11 papers (table 1).
To identify iciHHV-6 sequences in the SRA database, we developed a strategy to circumvent the prohibitively long download and search time it would take to examine thousands of records. First, we downloaded a list of 213,740 SRA accession numbers (chosen using NCBI Entrez; filtering for publicly accessible human DNA records). We then used a custom python script to web-scrape information from the SRA browser for each record. Specifically, we wanted to identify records for which a basic taxonomic assignment for the reads had been performed by NCBI’s in-house algorithm, the SRA Taxonomy Analysis Tool. The taxonomy information was not available for the majority of records. In cases where the analysis results were available, we considered any records where reads were tagged as “Roseolovirus.” To minimize the detection of false positives, we calculated an approximate read coverage for reads tagged as HHV-6 to compare with the coverage of human reads. In the cases of true iciHHV-6, we would expect a 1:2 ratio, but we examined all hits with a ratio between 1 and 10 to avoid false negatives since both the taxonomic analysis and initial sequencing are inexact procedures.
According to the above-described criteria, we downloaded a list of 97 records for mapping to an HHV-6A or HHV-6B reference genome and eliminated low-coverage records. After consolidating any of the remaining records that corresponded to the same sample (e.g., multiple runs), and excluding previously identified iciHHV-6 samples, we were left with a list of seven undescribed iciHHV-6 sequences that could be assembled into near full-length genomes, which was performed using SPAdes (Nurk et al. 2013).
Sample Collection, NGS Sequencing, and Phylogenetics
Patients in the heart disease cohort underwent a first endomyocardial biopsy (EMB) at the Institut Kardiale Diagnostik und Therapie in Berlin after excluding coronary artery disease. All patients presented with unexplained clinical symptoms of heart failure including fatigue, weakness, chest pain at rest or on exertion, dyspnea on exertion, palpitations and reduced physical capacity, and clinically suspected myocarditis or idiopathic dilated cardiomyopathy. All patients gave written informed consent for biopsy-based and genetic analyses to determine the underlying cause of the disease. The protocol was approved by the local medical ethics committee from the Charitè University Hospital Berlin, Germany. They underwent EMB and right heart catheterization in a standardized manner as previously described (Kühl et al. 2008).
The samples in the preeclampsia study are from the pregnancy outcome prediction study—a prospective cohort study of unselected nulliparous women with a singleton pregnancy attending the Rosie Hospital (Cambridge, UK) as previously described (Pasupathy et al. 2008; Sovio et al. 2015; Gaccioli et al. 2017). At 20 weeks of gestational age, maternal blood and paternal saliva were obtained for genotyping the parents. At the time of delivery, the placenta was systematically biopsied and a sample of umbilical cord was obtained for genotyping the offspring. We identified the iciHHV-6 positive samples by identifying a 1:1 ratio of the virus and a human housekeeping gene (RPP30) and used target enrichment and deep sequencing. We used droplet digital PCR to discriminate between HHV-6A and B and to select probes for the hybrid capture as described previously (Sedlak et al. 2014; Tweedy et al. 2015). To ensure that the samples are indeed from an iciHHV-6 patient, HHV-6A, HHV-6B, and the human RPP30 were quantified using specific primer and probes (Sedlak et al. 2014).
Approximately 100 ng of extracted gDNA was used to make sequencing libraries, as described previously (Greninger, Knudsen, et al. 2018; Greninger, Roychoudhury, Xie, et al. 2018). DNA was fragmented using the Kapa HyperPlus Kit (Roche) or the Covaris system followed by ligation of dual-indexed Truseq adapters. xGen Hybridization Capture reagents with either HHV-6B or HHV-6A/B custom capture probe pools (IDT and SureSelect) were used to enrich HHV-6, following manufacturer protocols. Sequencing was performed using 2×300 bp runs on an Illumina MiSeq. Genome assemblies for the heart disease cohort were created using a custom HHV-6 genome pipeline (https://github.com/proychou/HHV6, last accessed August 05, 2020). Sequencing reads and assemblies are available on GenBank with accession numbers MT508913–MT508970.
In the case of the preeclampsia cohort samples, Quality control and mapping was performed as part of the Pathogen Genomics Unit (PGU), Cambridge bioinformatics service. Quality filtering (base quality <30) and trimming was performed with TrimGalore (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/, last accessed August 05, 2020). Data were then mapped with Bbmap (https://jgi.doe.gov/data-and-tools/bbtools/bb-tools-user-guide/bbmap-guide/, last accessed August 05, 2020) to both HHV-6A/B and the best reference was selected based on coverage (references accessions NC_001664.4 and NC_000898.1, respectively). (A summary of mapping statistics can be found in supplementary table S2, Supplementary Material online and a representative example of mapped reads visualized in IGV is shown in supplementary fig. S3, Supplementary Material online) Bam files were then sorted and indexed with Samtools and duplicates removal performed with Picard. The reads were then assembled de novo using SPAdes (Nurk et al. 2013) and the chosen reference used to aid in scaffolding using PROmer (Kurtz et al. 2004). Although HHV6-A and HHV-6B are capable of coinfection (Leibovitch et al. 2014), we did not observe any evidence of samples containing both viral species. Although the genomes are extremely similar, there are nonetheless distinguishing SNPs which would have appeared as two separate populations of reads mapping to either reference virus. Moreover, because the phylogenetic separation between HHV-6A and HHV-6B is clear, a chimeric assembly would have resolved near the base of the tree which did not occur for any of our samples.
A total of 261 genome sequences of HHV-6A and 6B were aligned using a combination of MUSCLE (Edgar 2004) and manual adjustment. Each coding sequences was then extracted from the alignment and corrected for orientation and adjusted to maintain the correct reading frame, removing introns. The genes based on the refseq version NC_001664.4 of HHV-6A were concatenated and only the third-codon position was retained for phylogenetic reconstruction in MrBayes (Ronquist and Huelsenbeck 2003; Altekar et al. 2004), since the alignment length was well beyond the maximum allowed by the software. Each gene was designated a separate partition for the estimation of model parameters and the MCMC chains were run for 20 million generations, sampling every 1,000 generations. In addition to the Bayesian tree, we also reconstructed a phylogeny using the maximum Likelihood approach in RaxML, using the full sequence since the software does not have a length limitation (supplementary figs. S1 and S2, Supplementary Material online).
Fluorescence In Situ Hybridization
The HHV-6A/B genome and specific human chromosomes were detected by FISH as described previously (Kaufer 2013; Prusty et al. 2013; Wallaschek et al. 2016), with the following modifications and additions. HHV-6 probes were generated from HHV-6 BAC (strain U1102) and labeled using Biotin-High Prime (Sigma-Aldrich, St. Louis, MO); chromosomes probes were generated from chromosome-specific human BACs (clones RPCI-11; Source BioScience, Nottingham, England) and labeled using DIG-High prime (Sigma-Aldrich). Detection of probes signal was achieved using Cy3-Streptavidin for HHV-6 probes (1:200; Roche, Basel, Switzerland) and anti-DIG FITC Fab fragments for chromosomes probes (1:1,000; GE healthcare, Chicaco, IL). To obtain an adequate number of metaphases, the treatment of the cells with Colcemid for 16–24 h prior to preparation. DNA was stained with DAPI for 10 min (1:3,000; Biolegend, San Diego, CA), followed by washes in 1× PBS. Slides were mounted with a drop of ProLong Glass Antifade Mountant (ThermoFisher, Waltham, MA). Images were acquired with a Zeiss M1 Microscope using a 100× objective and Axio Vision software (Carl Zeiss, Inc). Images were analyzed using ImageJ (https://imagej.nih.gov/ij/, last accessed August 05, 2020) and its specific processing package Fiji (https://imagej.net/Fiji, last accessed August 05, 2020).
Dating Analysis
We performed a simple estimate of the divergence times of clades of endogenous HHV-6 that we propose originate from a single integration event. This assumes that SNPs observed in the viral genomes of each clade accumulated after endogenization, and therefore can be used as a measure of age when combined with an appropriate mutation rate. The age of a clade was calculated , where T is the time before present, D is the mean pairwise distance of iciHHV-6 genomes in a clade, n is the number of tips in a clade, and R is the average number of mutation/site/year. This basic approach is similar to the method commonly used to estimate integration dates for endogenous retroviruses (Hayward 2017). We employed two different rate estimates (0.5× and 1×), reviewed in Scally and Durbin (2012), since we do not have a direct measure of the rate for endogenous HHV 6. The rate of 0.5× is based on observations of per-generation mutations in modern humans and assumes a generation time of between 20 and 30 years. Another rate that is often used is 1×, which is derived from the phylogenetic divergence between humans and other great apes or old-world monkeys. Because iciHHV-6 may accumulate mutations at a higher rate than the rest of the genome, we also used R = 2× to illustrate such a scenario on the age estimate. We also recalculated the times of integration using the upper and lower limits of the 95% confidence interval for the distance estimates.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We are very grateful to the editorial team and anonymous reviewers for contributing to the improvement of this article. We are grateful to Ann Reum and Annett Neubert for their technical assistance, and to Dan Depledge (UCL) for designing the baits used for the sequencing of the preeclampsia study samples and to Rachel Williams for performing the sequencing. We are also grateful for the advice and help of Dr. Loris Bennett and the rest of the HPC team at the scientific computing unit of the Free University Berlin.
This study was supported by the Einstein International Postdoctoral Award EIPF-Aswad and the ERC starting grant Stg 677673 awarded to A.A. and B.B.K., respectively. J.B. receives funding from the NIHR UCL/UCLH BRC. This work was also supported by the Women’s Health theme of the NIHR Cambridge Biomedical Research Centre and the Medical Research Council (MR/K021133/1 and G1100221 awarded to G.C.S.S. and D.S.C.-J.). We also thank the HHV-6 foundation for reagent support.
References
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F.. 2004. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics 20(3):407–415. [DOI] [PubMed] [Google Scholar]
- Aswad A, Katzourakis A.. 2014. The first endogenous herpesvirus, identified in the tarsier genome, and novel sequences from primate rhadinoviruses and lymphocryptoviruses. PLoS Genet. 10(6):e1004332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aswad A, Katzourakis A.. 2016. Paleovirology: the study of endogenous viral elements. In: Weaver SC, Denison M, Roossinck M, Vignuzzi M, editors. 1st ed. Virus evolution: current research and future directions. Poole: Caister Academic Press. p. 273–292. [Google Scholar]
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, et al. 2020. Insights into human genetic variation and population history from 929 diverse genomes. Science 367(6484):eaay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JR, Roy S, Ruis C, Yara Romero E, Shah D, Williams R, Breuer J.. 2016. Norovirus whole-genome sequencing by SureSelect target enrichment: a robust and sensitive method. J Clin Microbiol. 54(10):2530–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-Duque J-C, Adhikari K, Fuentes-Guajardo M, Mendoza-Revilla J, Acuña-Alonzo V, Barquera R, Quinto-Sánchez M, Gómez-Valdés J, Everardo Martínez P, Villamil-Ramírez H, et al. 2018. Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nat Commun. 9(1):5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BB. 2015. A neonate with acute heart failure: chromosomally integrated human herpesvirus 6–associated dilated cardiomyopathy. J Pediatr. 167(1):188–192.e1. [DOI] [PubMed] [Google Scholar]
- Depledge DP, Palser AL, Watson SJ, Lai IY-C, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J.. 2011. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One 6(11):e27805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez G, Dambaugh TR, Stamey FR, Dewhurst S, Inoue N, Pellett PE.. 1999. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 73(10):8040–8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A, Watanabe K, Ohye T, Suzuki K, Matsubara T, Shimizu N, Kurahashi H, Yoshikawa T, Katano H, Inoue N, et al. 2014. Molecular and virological evidence of viral activation from chromosomally integrated human herpesvirus 6A in a patient with X-linked severe combined immunodeficiency. Clin Infect Dis. 59(4):545–548. [DOI] [PubMed] [Google Scholar]
- Gaccioli F, Lager S, Sovio U, Charnock-Jones DS, Smith GCS.. 2017. The pregnancy outcome prediction (POP) study: investigating the relationship between serial prenatal ultrasonography, biomarkers, placental phenotype and adverse pregnancy outcomes. Placenta 59(Suppl 1):S17–S25. [Google Scholar]
- Gompels UA, Nicholas J, Lawrence G, Jones M, Thomson BJ, Martin MED, Efstathiou S, Craxton M, Macaulay HA.. 1995. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology 209(1):29–51. [DOI] [PubMed] [Google Scholar]
- Gravel A, Ablashi D, Flamand L. 2013. Complete genome sequence of early passaged human herpesvirus 6A (GS strain) isolated from North America. Genome Announc. 1(3):e00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel A, Dubuc I, Morissette G, Sedlak RH, Jerome KR, Flamand L.. 2015. Inherited chromosomally integrated human herpesvirus 6 as a predisposing risk factor for the development of angina pectoris. Proc Natl Acad Sci U S A. 112(26):8058–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel A, Hall CB, Flamand L.. 2013. Sequence analysis of transplacentally acquired human herpesvirus 6 DNA is consistent with transmission of a chromosomally integrated reactivated virus. J Infect Dis. 207(10):1585–1589. [DOI] [PubMed] [Google Scholar]
- Greninger AL, Knudsen GM, Roychoudhury P, Hanson DJ, Sedlak RH, Xie H, Guan J, Nguyen T, Peddu V, Boeckh M, et al. 2018. Comparative genomic, transcriptomic, and proteomic reannotation of human herpesvirus 6. BMC Genomics 19(1):204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Roychoudhury P, Makhsous N, Hanson D, Chase J, Krueger G, Xie H, Huang M-L, Saunders L, Ablashi DV, et al. 2018. Copy number heterogeneity, large origin tandem repeats, and interspecies recombination in human herpesvirus 6A (HHV-6A) and HHV-6B reference strains. J Virol. 92(10):e00135–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greninger AL, Roychoudhury P, Xie H, Casto A, Cent A, Pepper G, Koelle DM, Huang M-L, Wald A, Johnston C, et al. 2018. Ultrasensitive capture of human herpes simplex virus genomes directly from clinical samples reveals extraordinarily limited evolution in cell culture. mSphere 3(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CB, Caserta MT, Schnabel KC, Shelley LM, Carnahan JA, Marino AS, Yoo C, Lofthus GK.. 2010. Transplacental congenital human herpesvirus 6 infection caused by maternal chromosomally integrated virus. J Infect Dis. 201(4):505–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CB, Long CE, Schnabel KC, Caserta MT, McIntyre KM, Costanzo MA, Knott A, Dewhurst S, Insel RA, Epstein LG.. 1994. Human herpesvirus-6 infection in children – a prospective study of complications and reactivation. N Engl J Med. 331(7):432–438. [DOI] [PubMed] [Google Scholar]
- Hayward A. 2017. Origin of the retroviruses: when, where, and how? Curr Opin Virol. 25:23–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, HallSedlak R, Magaret A, Huang M-L, Zerr DM, Jerome KR, Boeckh M.. 2016. Efficient identification of inherited chromosomally integrated human herpesvirus 6 using specimen pooling. J Clin Virol. 77:71–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Magaret AS, Hall-Sedlak R, Mikhaylova A, Huang M-L, Sandmaier BM, Hansen JA, Jerome KR, Zerr DM, Boeckh M.. 2017. Outcomes of hematopoietic cell transplantation using donors or recipients with inherited chromosomally integrated HHV-6. Blood 130(8):1062–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Zerr DM.. 2014. Roseoloviruses in transplant recipients: clinical consequences and prospects for treatment and prevention trials. Curr Opin Virol. 9:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Hidalgo-Bravo A, Zhang E, Cotton VE, Mendez-Bermudez A, Wig G, Medina-Calzada Z, Neumann R, Jeffreys AJ, Winney B, et al. 2014. Human telomeres that carry an integrated copy of human herpesvirus 6 are often short and unstable, facilitating release of the viral genome from the chromosome. Nucleic Acids Res. 42(1):315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, et al. 1999. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol. 73(10):8053–8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ.. 2010. Endogenous viral elements in animal genomes. PLoS Genet. 6(11):e1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Gifford RJ, Tristem M, Gilbert MTP, Pybus OG.. 2009. Macroevolution of complex retroviruses. Science 325(5947):1512–1512. [DOI] [PubMed] [Google Scholar]
- Kaufer B. 2013. Detection of integrated herpesvirus genomes by fluorescence in situ hybridization (FISH). Virus Host Interact Methods Protoc. 2013;1064:141–152. [DOI] [PubMed] [Google Scholar]
- Kaufer B, Flamand L.. 2014. Chromosomally integrated HHV-6: impact on virus, cell and organismal biology. Curr Opin Virol. 2014;9:111–118. [DOI] [PubMed] [Google Scholar]
- Kühl U, Lassner D, Pauschinger M, Gross UM, Seeberg B, Noutsias M, Poller W, Schultheiss H-P.. 2008. Prevalence of erythrovirus genotypes in the myocardium of patients with dilated cardiomyopathy. J Med Virol. 80(7):1243–1251. [DOI] [PubMed] [Google Scholar]
- Kühl U, Lassner D, Wallaschek N, Gross UM, Krueger GRF, Seeberg B, Kaufer BB, Escher F, Poller W, Schultheiss H-P.. 2015. Chromosomally integrated human herpesvirus 6 in heart failure: prevalence and treatment. Eur J Heart Fail. 17(1):9–19. [DOI] [PubMed] [Google Scholar]
- Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL.. 2004. Versatile and open software for comparing large genomes. Genome Biol. 5(2):R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitch EC, Brunetto GS, Caruso B, Fenton K, Ohayon J, Reich DS, Jacobson S.. 2014. Coinfection of human herpesviruses 6A (HHV-6A) and HHV-6B as demonstrated by novel digital droplet PCR assay. PLoS One 9(3):e92328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Lin L, Malhotra R, Yang L, Acharya R, Poss M.. 2019. A computational framework to assess genome-wide distribution of polymorphic human endogenous retrovirus-K In human populations. PLOS Comput Biol. 15(3):e1006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadpour Touserkani F, Gaínza-Lein M, Jafarpour S, Brinegar K, Kapur K, Loddenkemper T.. 2017. HHV-6 and seizure: a systematic review and meta-analysis. J Med Virol. 89(1):161–169. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Akey JM, Jakobsson M, Pritchard JK, Tishkoff S, Willerslev E.. 2017. Tracing the peopling of the world through genomics. Nature 541(7637):302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, et al. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads. Heidelberg (Berlin): Springer. p. 158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N, Wallaschek N, Kaufer BB.. 2014. Herpesvirus genome integration into telomeric repeats of host cell chromosomes. Annu Rev Virol. 1(1):215–235. [DOI] [PubMed] [Google Scholar]
- Pantry SN, Medveczky MM, Arbuckle JH, Luka J, Montoya JG, Hu J, Renne R, Peterson D, Pritchett JC, Ablashi DV, et al. 2013. Persistent human herpesvirus-6 infection in patients with an inherited form of the virus. J Med Virol. 85(11):1940–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasupathy D, Dacey A, Cook E, Charnock-Jones DS, White IR, Smith GCS.. 2008. Study protocol. A prospective cohort study of unselected primiparous women: the pregnancy outcome prediction study. BMC Pregnancy Childbirth 8(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellett PE, Ablashi DV, Ambros PF, Agut H, Caserta MT, Descamps V, Flamand L, Gautheret-Dejean A, Hall CB, Kamble RT, et al. 2012. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol. 22(3):144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton TJ, Wang C, Li JZ, Rosenberg NA.. 2010. Inference of unexpected genetic relatedness among individuals in HapMap Phase III. Am J Hum Genet. 87(4):457–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusty BK, Krohne G, Rudel T, Tanaka-Taya K, Sashihara J, Kurahashi H, Amo K, Miyagawa H, Arbuckle J, Medveczky M, et al. 2013. Reactivation of chromosomally integrated human herpesvirus-6 by telomeric circle formation. PLoS Genet. 9(12):e1004033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP.. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574. [DOI] [PubMed] [Google Scholar]
- Scally A, Durbin R.. 2012. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 13(10):745–753. [DOI] [PubMed] [Google Scholar]
- Sedlak RH, Cook L, Huang M-L, Magaret A, Zerr DM, Boeckh M, Jerome KR.. 2014. Identification of chromosomally integrated human herpesvirus 6 by droplet digital PCR. Clin Chem. 60(5):765–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovio U, White IR, Dacey A, Pasupathy D, Smith GCS.. 2015. Screening for fetal growth restriction with universal third trimester ultrasonography in nulliparous women in the Pregnancy Outcome Prediction (POP) study: a prospective cohort study. Lancet 386(10008):2089–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant PH, Rausch T, Gardner EJ, Handsaker RE, Abyzov A, Huddleston J, Zhang Y, Ye K, Jun G, Fritz MH-Y, et al. 2015. An integrated map of structural variation in 2,504 human genomes. Nature 526(7571):75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarlinton RE, Meers J, Young PR.. 2006. Retroviral invasion of the koala genome. Nature 442(7098):79–81. [DOI] [PubMed] [Google Scholar]
- Telford M, Navarro A, Santpere G.. 2018. Whole genome diversity of inherited chromosomally integrated HHV-6 derived from healthy individuals of diverse geographic origin. Sci Rep. 8(1):3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedy J, Spyrou MA, Donaldson CD, Depledge D, Breuer J, Gompels UA.. 2015. Complete genome sequence of the human herpesvirus 6A strain AJ from Africa resembles strain GS from North America. Genome Announc. 3(1):e01498–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedy JG, Spyrou MA, Pearson M, Lassner D, Kuhl U, Gompels UA.. 2016. Complete genome sequence of germline chromosomally integrated human herpesvirus 6A and analyses integration sites define a new human endogenous virus with potential to reactivate as an emerging infection. Viruses 8(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallaschek N, Sanyal A, Pirzer F, Gravel A, Mori Y, Flamand L, Kaufer BB.. 2016. The telomeric repeats of human herpesvirus 6A (HHV-6A) are required for efficient virus integration. PLOS Pathog. 12(5):e1005666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildschutte JH, Williams ZH, Montesion M, Subramanian RP, Kidd JM, Coffin JM.. 2016. Discovery of unfixed endogenous retrovirus insertions in diverse human populations. Proc Natl Acad Sci U S A. 113(16):E2326–E2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E, Bell AJ, Wilkie GS, Suárez NM, Batini C, Veal CD, Armendáriz-Castillo I, Neumann R, Cotton VE, Huang Y, et al. 2017. Inherited chromosomally integrated human herpesvirus 6 genomes are ancient, intact, and potentially able to reactivate from telomeres. J Virol. 91(22):JVI.01137-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ECotton VEHidalgo-Bravo AHuang YBell AJJarrett RFWilkie GSDavison AJNacheva EPSiebert R,et al. . 2016. HHV-8-unrelated primary effusion-like lymphoma associated with clonal loss of inherited chromosomally-integrated human herpesvirus-6A from the telomere of chromosome 19q. Sci Rep. 6:22730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.