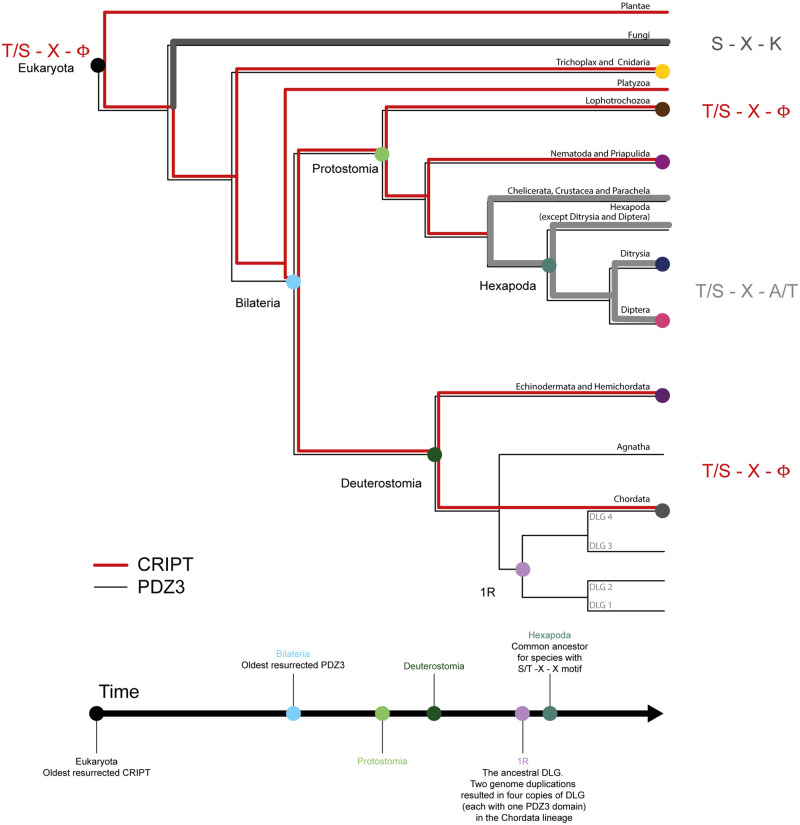
Fig. 2.
Evolution of DLG PDZ3 and the PDZ-binding motif in CRIPT. Simplified tree of organismal groups used for ancestral sequence reconstructions of CRIPT and PDZ3. The nodes in the phylogenetic tree are highlighted in the color code used throughout the article for extant and reconstructed ancestral CRIPT and PDZ3 domain variants. Evolution of CRIPT C-terminal-binding motif is highlighted by red (type I: T/S–X–Φ) to gray (undefined motif). Ancestral sequence reconstructions of CRIPT and PDZ3, respectively, were performed based on multiple sequence alignments and a species tree (CommonTree and OpenTree). In the reconstruction, 309 unique sequences of CRIPT from 498 different species and 249 unique sequences of PDZ3 from the DLG protein family from 324 different species were included.