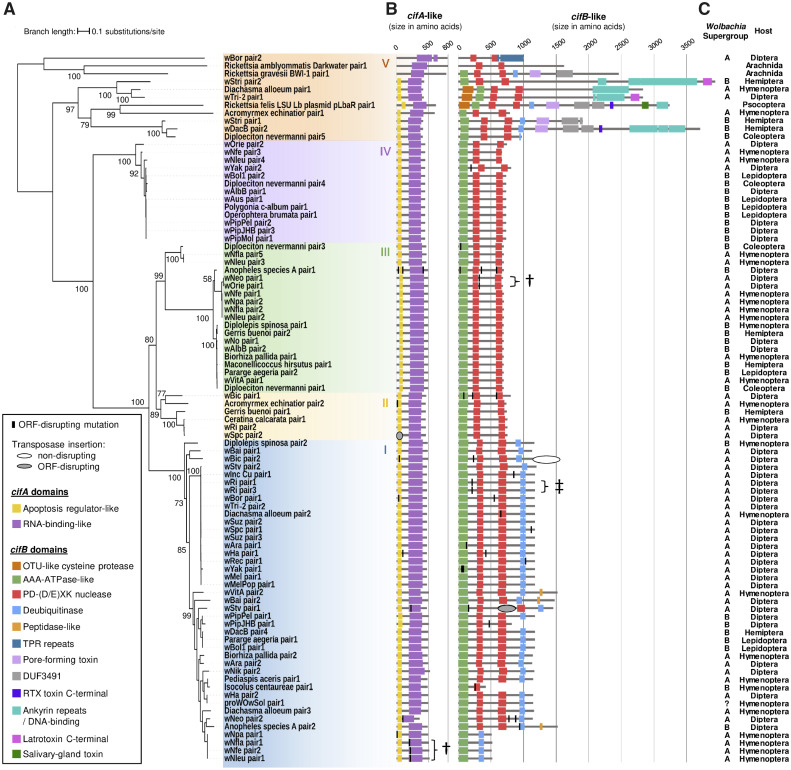
Fig. 2.
Phylogeny and protein domain architecture of cifA and cifB. (A) Maximum likelihood tree of concatenated cifA and cifB nucleotide sequences. Partially sequenced cif homologs were excluded. The tree is midpoint rooted. Bootstrap values were estimated from 1,000 replicates. (B) Protein domains. Mutations that disrupt the ORF are indicated by a vertical bar. Symbols indicate where the same ORF-disrupting mutation is found in two homologs due to speciation (†) or duplication (‡) events. All domains had a HHpred probability of being true positives >75% in at least one sequence. The suffix “-like” at the end of the domain name indicates that there were no sequences where the probability of the domain being true was >95%. Details of the domains are in supplementary table S5, Supplementary Material online. (C) Wolbachia supergroups and arthropod hosts from which the cif sequences were isolated.