Amoebae are protists that have complicated relationships with bacteria, covering the whole spectrum of symbiosis. Amoeba-bacterium interactions contribute to the study of predation, symbiosis, pathogenesis, and human health. Given the complexity of their relationships, it is necessary to understand the ecology and evolution of their interactions. In this paper, we provide an updated review of the current understanding of amoeba-bacterium interactions. We start by discussing the diversity of amoebae and their bacterial partners.
KEYWORDS: amoebae, bacteria, protist, ecology, evolution, mutualism, parasitism, symbiosis
ABSTRACT
Amoebae are protists that have complicated relationships with bacteria, covering the whole spectrum of symbiosis. Amoeba-bacterium interactions contribute to the study of predation, symbiosis, pathogenesis, and human health. Given the complexity of their relationships, it is necessary to understand the ecology and evolution of their interactions. In this paper, we provide an updated review of the current understanding of amoeba-bacterium interactions. We start by discussing the diversity of amoebae and their bacterial partners. We also define three types of ecological interactions between amoebae and bacteria and discuss their different outcomes. Finally, we focus on the implications of amoeba-bacterium interactions on human health, horizontal gene transfer, drinking water safety, and the evolution of symbiosis. In conclusion, amoeba-bacterium interactions are excellent model systems to investigate a wide range of scientific questions. Future studies should utilize advanced techniques to address research gaps, such as detecting hidden diversity, lack of amoeba genomes, and the impacts of amoeba predation on the microbiome.
WHAT ARE AMOEBAE?
Amoebae are protists that move using pseudopods and feed by phagocytosis. They are widespread and mostly found in water, soil, and air. A German naturalist, August Johann Rösel von Rosenhof, recorded the first known amoeba in 1755, which is similar to Amoeba proteus according to his illustrations. Traditional classification placed amoebae in the Sarcodina group based on morphology and further divided them into Rhizopodea and Actinopodea, depending on the type of pseudopodia (1). Such classification was widely accepted and used because it was convenient, and no alternative system existed at that time (2). Many East Asian translations, such as Chinese, still use it.
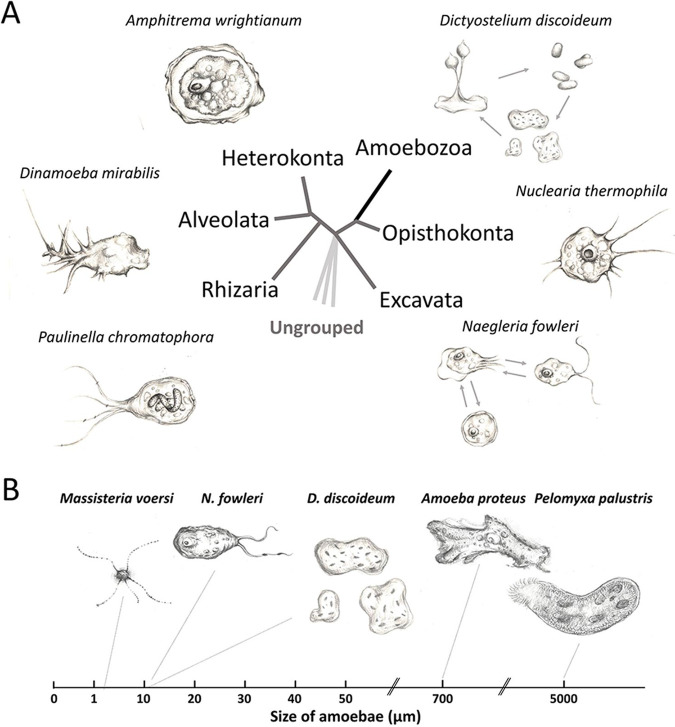
However, molecular phylogenetic studies using marker genes such as the 18S rRNA have shown that Sarcodina is not a monophyletic group. Instead, they are distributed widely across the eukaryote phylogeny and show great diversity, including Amoebozoa, Rhizaria, Excavata, Heterokonta, Alveolata, Opisthokonta, and other ungrouped species (3) (Fig. 1, Table 1). Among them, Amoebozoa is the only group that solely consists of amoebae, and it is also the most diverse group, containing over 17,000 species that differ in their lifestyles (free-living versus parasitic) (4, 5). Other groups of amoebae also show great diversity. For instance, Rhizaria contains filose amoebae, whose filose pseudopods are narrow and tapering, and all those that produce shells. The Heterolobosea in the Excavata supergroups include amoebae that can transform between amoeboid and flagellate forms (4). Amoebae span a wide range of sizes, ranging from several to thousands of micrometers, and their size mostly determines their predation (Fig. 1B). For instance, in testate amoebae, the prey size is limited by their shell size (6). In general, different amoebae feed on bacteria, algae, flagellates, and ciliates depending on their sizes.
FIG 1.
Schematic representation of amoeba diversity. (A) Amoebae are spread in several supergroups, including Amoebozoa, Rhizaria, Excavata, Heterokonta, Alveolata, Opisthokonta, and other ungrouped species. Amoebozoa (black) is the only group that solely consists of amoebae. The tree topology is from previous classifications (3, 139). (B) Amoebae show considerable variation in their sizes.
TABLE 1.
List of nonpredatory interactions between amoebae and bacteria
| Supergroups and major genera | Bacteria | Reference |
|---|---|---|
| Amoebozoa | ||
| Acanthamoeba | Caedibacter, “Jidaibacter,” “Odysella,” “Paracaedibacter,” Procabacter, “Berkiella,” Pseudomonas, Stenotrophomonas, Flavobacterium, Amoebophilus, Neochlamydia, Parachlamydia, Protochlamydia, Listeria, Mycobacterium, Afipia, Ralstonia, Rickettsia, Simkania, Salmonella, Yersinia, Campylobacter, Arcobacter, Staphylococcus, Mobiluncus, Bacillus, Burkholderia, Escherichia, Helicobacter, Porphyromonas, Prevotella, Chlamydophila, Waddlia, Coxiella, Francisella | 7, 13, 14, 16, 136, 148 |
| Dictyostelium | Legionella, Escherichia, Klebsiella, Paraburkholderia, Pseudomonas, Mycobacterium, Bordetella, Salmonella | 18, 19, 21–23, 99, 101, 102, 149, 150 |
| Hartmannella | “Nucleicultrix,” Legionella, Pseudomonas, Simkania, Neochlamydia, Bacillus | 7, 14, 151 |
| Echinamoebae | Pseudomonas, Caulobacter | 14, 84 |
| Balamuthia | Simkania | 152 |
| Vannellae | “Occultobacter,” “Mesochlamydia” | 153, 154 |
| Saccamoebae | “Mesochlamydia,” Ehrlichia | 154, 155 |
| Cochliopodium | “Cochliophilus” | 156 |
| Pelomyxa | Syntrophorhabdus, Rhodococcus | 157 |
| Entamoeba | Escherichia | 44 |
| Amoeba | Legionella | 158 |
| Chaos | ||
| Thecamoebae | ||
| Rhizaria | ||
| Paulinella | “Plastid” | |
| Gyromitus | ||
| Vampyrella | ||
| Excavata | ||
| Naegleria | Stenotrophomonas, Legionella, Protochlamydia, Simkania, Neochlamydia, Acidovorax, Flavobacterium | 14, 120, 136 |
| Sawyeria | ||
| Psalteriomonas | Methanobacterium | 159 |
| Vahlkampfia | ||
| Stygamoeba | ||
| Dientamoeba | ||
| Heteramoeba | ||
| Tulamoeba | ||
| Heterokonta | ||
| Chrysamoeba | ||
| Opisthokonta | ||
| Nuclearia | Planktothrix, Ovatusbacter | 160, 161 |
| Micronuclearia | ||
| Alveolata | ||
| Dinamoeba | ||
| Pfiesteria | ||
| Others | ||
| Astramoeba | ||
| Dientamoeba | ||
| Malamoeba |
Amoebae are essential components of aquatic and terrestrial ecosystems and play a vital role in the dynamics of microbial communities, nutrient cycling, and energy flow (7, 8). They are also a potential threat to human health, as some of them are pathogenic or even lethal to humans (7, 9, 10). There are several excellent reviews on amoebae and their bacterial symbionts (11–15). In recent years, amoeba-bacterium interactions have attracted increasing interest in a wide range of disciplines, including molecular and cellular biology, medicine, environmental science, ecology, and evolution (16–25). Therefore, we want to provide an updated review of the current understanding of amoeba-bacterium interactions, with a particular focus on the ecological and evolutionary aspects. We identify key research areas and gaps and highlight how ecological-evolutionary interaction could increase our understanding of the role of amoeba-bacterium interactions in both nature and the laboratory.
WHY DO AMOEBA-BACTERIUM INTERACTIONS MATTER?
Long before bacteria interacted with animals and humans, they interacted with amoebae (9). Therefore, it is not surprising that amoeba-bacterium interactions are complex and cover the whole range of symbiosis (9). Amoebae have evolved different mechanisms to find, kill, and digest bacteria, while bacteria also developed strategies to resist amoeba predation and, in return, sometimes to infect and kill amoebae.
We can learn a lot from the long-term coevolution between amoebae and bacteria. First, it gives us a unique chance to study bacterium-eukaryote interactions. A key challenge in ecology and evolutionary biology is to disentangle the relationship and coevolution between the eukaryotic host and its associated bacteria, which often requires the recombination of different partners. However, it is challenging to separate, mix, and match the host with its bacteria using existing model systems, which limits our ability to understand the interaction and coevolution between them. Amoeba-bacterium interactions can provide a simple and tractable model system to address these issues (19–21, 26–28). Second, it gives us a model system to investigate how intracellular endosymbiosis evolves. Intracellular endosymbionts have evolved specialized lifestyles to survive within the host cells (29). The transition from free-living to intracellular lifestyles is often accompanied by genome reduction (30, 31). Many isolated bacteria, such as Rickettsia and Wolbachia species, showed a significant genome reduction in amoebae. Further analysis suggested that amoebae serve as a melting pot that allows diverse bacteria to adapt to the intracellular lifestyle or create new pathogens (11, 32). Finally, it can provide some insights into human pathogens and diseases. Amoebae can also serve as environmental reservoirs for many human pathogens (11, 12, 14). In addition, many of the core mechanisms used by amoebae to ingest and kill bacteria have been evolutionarily conserved in human phagocytic cells (33, 34). Therefore, understanding the evolution of amoeba-bacterium interactions will also increase our understanding of amoebae’s role in the origin, spread, and control of infectious diseases.
HOW DO AMOEBAE INTERACT WITH BACTERIA?
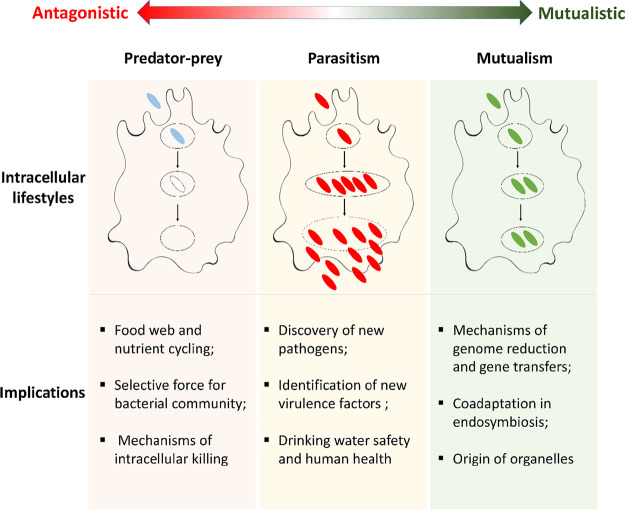
Amoeba-bacterium interactions cover the whole spectrum of symbiotic relationships, ranging from antagonism to mutualism. In this review, we mainly discuss three types of ecological interactions (Fig. 2): predation, parasitism, and mutualism. However, from an evolutionary view, it is essential to note that these interactions are continuous rather than discrete, and different interactions can evolve from one to another under selective pressures (Fig. 2).
FIG 2.
Diagram of ecological interactions between amoebae and bacteria. The figure represents three general types of amoeba-bacterium interactions as well as their impacts. Bacteria can have different interactions with amoebae, ranging from antagonism to mutualism. Their interactions also can move from one category to another, making this a great system to investigate how antagonistic interactions evolve to be more mutualistic. Blue, bacterial prey. Red, pathogenic bacteria that can replicate and escape to the cytosol and the environment. Green, endosymbionts of amoebae.
Predation.
Predation is the dominant relationship that amoebae have with bacteria. Amoeba predation can be a powerful structuring force for the natural bacterial community (35). Protist consumption, including by amoebae, is a significant source of bacterial mortality in most aquatic and terrestrial ecosystems (36). Bacteria should often be under strong selection pressure to evade predation and deploy a wide range of chemicals that serve that purpose. Resistance to predation may sometimes be the first step toward pathogenicity and virulence, not just toward predators but to other eukaryotes (37). Nevertheless, amoebae can consume a vast range of bacteria. The social amoeba Dictyostelium discoideum grows on the majority of bacteria tested (38, 39). For example, in a study of 159 bacterial isolates collected from soil and fecal samples containing D. discoideum, 123 supported amoeba growth and development to some degree (38). The majority were Proteobacteria (alpha, beta, and gamma), but others were taxonomically distributed in the Bacteroidetes, Firmicutes, and Actinobacteria. Similarly, other Dictyostelids have shown generalist abilities (40, 41) as have, although less thoroughly studied, other Amoebozoans (42–44).
The degree of generalism may be even higher, because some inedible bacteria become edible when they are at lower densities (37), as might be expected if there are defensive compounds that get diluted. Similarly, D. discoideum’s own secretions make it a more effective predator when it is at higher densities (45). Arguably, amoebae may show a degree of diet generalism far greater than better-known generalists, like birds, which can eat a wide variety of arthropods. Therefore, amoebae may provide an interesting system for studying how generalism evolves. Supergeneralist predation raises the question of how these predators cope with the defenses of so many prey or, alternatively, why so many prey seem defenseless. The answer is unknown, but D. discoideum does show quite different transcriptional profiles on different bacteria, suggesting that it is adjusting its feeding strategies to different prey (46, 47).
Understanding how amoebae differentially impact bacteria also is essential to evaluating their contribution to ecosystem processes. However, we know very little about the mechanisms of selective grazing behavior of amoebae. Below we will discuss the two significant steps of amoeba predation.
(i) Recognition of food bacteria. Recognition of food bacteria is the first step for a successful hunt. Theoretically, amoebae should be able to sense, recognize, and move to their food bacteria. They should also need to distinguish and avoid pathogenic bacteria. However, these are hypotheses that need experimental testing. Several studies reported that the social amoeba D. discoideum had different gene expression profiles when exposed to different bacteria (46–48), and a different set of genes were activated when they fed on Gram-positive and Gram-negative bacteria (46, 47). A more recent study reported that Dictyostelium amoebae respond in a highly specific manner to different bacteria species (47). They were also more attracted to Gram-negative bacteria in a chemotaxis assay (49). Nevertheless, among these differentially expressed genes, it is challenging to distinguish metabolic adaptation from bacterial sensing. Other studies have used random mutagenesis to discover amoeba genes essential for bacterial sensing, including fspA, tirA, and lrrkA (50–52). For instance, a D. discoideum amoeba mutant (fspA knockout) cannot grow on noncapsulated Klebsiella pneumoniae, but it grows well on capsulated LM21 K. pneumoniae, indicating that D. discoideum amoebae use the fspA gene to recognize K. pneumoniae capsules (51). Another study found that vps13F had a significant role in bacterial recognition and intracellular killing in D. discoideum (53). Taken together, these studies suggest that D. discoideum does have specific mechanisms to sense bacteria.
However, it is essential to note that most of the studies focused on D. discoideum amoebae, and we know very little about the sensing mechanisms in other groups of amoebae. One study compared the foraging strategies between two commonly used amoebae, Acanthamoeba and Dictyostelium (54). Surprisingly, they found that, unlike D. discoideum, Acanthamoeba did not use chemotaxis to sense its food bacteria. Given the vast diversity of amoebae, this raises the question of how variable the foraging strategies are across the major clades of amoebae.
(ii) Molecular mechanisms of intracellular killing. Amoebae are very effective bacterial killers (33). They track bacteria through chemotaxis and kill them using phagocytosis. Phagocytosis is the process by which cells ingest or engulf other cells or particles, and amoebae can engulf and kill most bacteria. After engulfment, the phagosome undergoes maturation, making it a highly acidic, degradative, and oxidative compartment (55). Amoebae use various strategies to kill bacteria. Phagosome acidification is the first step, in which V-ATPase plays a central role (56). In addition, the phagosome also contains proteases (57), hydrolases (58), certain metals (59, 60), and ROS (reactive oxygen species) (61), which combine to kill and break down bacteria (33, 60, 62). With all these bacterial control strategies, amoebae can effectively kill and digest bacteria.
Dozens of genes have been reported to play essential roles in the intracellular killing of amoebae using two different strategies. The first strategy is to find the orthologues of the mammalian system in amoeba genomes, which has successfully identified genes such as alyA (58), catD (57), lvsB (63), nox2 (61), nramp (64), and wshA (65) in D. discoideum. For example, nox2 is crucial for the oxidative burst of phagocytic cells and plays a significant role in intracellular killing in mammals (66). Three orthologues of nox2 have been identified in the D. discoideum genome (61). Another strategy is to use killing-deficient mutants to discover novel killing genes. Random screens of D. discoideum mutants have found that fspA (51), kil1 (48), kil2 (67), phg1a (68), and tirA (50) genes played an essential role in the intracellular killing. Still, we are just beginning to understand this complex mechanism. For instance, in D. discoideum phg1, kil1, and kil2 are essential for the intracellular killing of K. pneumoniae (a common food bacterium in the laboratory), but they are not necessary for Pseudomonas aeruginosa and Bacillus subtilis (33). This indicates that D. discoideum has independent mechanisms to kill different bacteria. Bacteria of the same species also can differ radically in edibility. Future studies should investigate the molecular mechanisms of intracellular killing in other groups of amoebae. Given the huge diversity across amoebae, an educated guess would be that many new intracellular killing genes will be identified.
Parasitic interactions.
Predation is a potent agent of natural selection; therefore, bacteria will inevitably evolve to adapt to it, and some even infect amoebae in return. These bacteria are called amoeba-resisting bacteria (ARBs) because they have evolved to become resistant to amoebae. ARBs can escape the internalization of amoeba feeding or can survive, replicate, and exit amoebae after internalization (39). ARBs that have evolved to infect amoebae are of particular interest to microbiologists and medical researchers because many are human pathogens.
(i) Legionella pneumophila. Among ARBs, L. pneumophila is one of the best-studied systems (69–73). It is ubiquitous and has evolved mechanisms to infect and replicate within amoebae and humans, causing pneumonia termed Legionnaires’ disease. Amoebae are the environmental reservoirs for L. pneumophila and play an essential role in human infection (74, 75). The adaptation and coevolution between L. pneumophila and amoebae have shaped L. pneumophila’s genome, resulting in a large number of effector proteins that play an essential role in its intracellular lifestyle in human macrophages (76). For example, L. pneumophila uses an F-box effector to exploit the farnesylation machinery of eukaryotic host cells (77). L. pneumophila also can promote the host’s proteasomal degradation for its benefit and can affect the ubiquitination pathway (69, 78, 79). More recently, a study found that L. pneumophila injects LamA, a Legionella amylase, into the cytosol of human macrophages and amoebae, which degrades host cell glycogen (80). LamA interferes with amoeba host-specific processes (subverts encystation of the amoebae) and triggers accidental inflammatory responses in humans (80). Finally, L. pneumophila can also gain protection against disinfection processes from amoebae (81). These results suggest that amoeba-Legionella is a useful model system that increases our understanding of the parasitic interactions between amoebae and pathogens.
(ii) Mycobacteria. Mycobacterium, a genus of Actinobacteria, contains pathogens such as Mycobacterium tuberculosis, M. ulcerans, M. leprae, and other nontuberculous mycobacteria (NTM). They can cause severe diseases such as tuberculosis and leprosy in humans. In nature, mycobacteria frequently have been isolated from amoebae in different habitats, including drinking water systems (82–84), hospitals (85), and cooling towers (86). Many studies try to disentangle the interactions between amoebae and mycobacteria. One study found that M. tuberculosis and M. marinum can escape from their amoeba hosts through nonlytic ejection (23). Interestingly, M. avium did not eject but remained within its amoeba host (23). The following study suggested that nonlytic transmission is dependent on the autophagy pathway (22). A recent study reported that Mycobacterium bovis, part of the M. tuberculosis complex, can evade D. discoideum predation through the ESX-1 type VII secretion system (87). This study also supports the hypothesis that amoebae are a training ground for pathogens.
NTM can also have complex interactions with amoebae. For instance, the cooccurrence of NTM and amoebae has been reported in hospital water networks, and Mycobacterium avium showed high replication rates only in Acanthamoeba lenticulata (85). Another study reported the association of NTM and amoebae in the drinking water network (82). It also has been reported that NTM smaller than 2 μm do not replicate intracellularly and do not kill amoebal trophozoites, indicating size-correlated relationships between NTM and their amoeba hosts, which implies NTM can interact differently with amoebae (88).
(iii) Chlamydiae. The Chlamydiae are a phylum of obligate intracellular bacteria. Some of them are nonpathogenic symbionts, while others are important human pathogens (89). For instance, Chlamydia trachomatis can cause sexually transmitted infections and eye disease trachoma (90). Chlamydia pneumoniae infection is related to arthritis, asthma, and atherosclerosis (91). Chlamydia has been frequently isolated from amoebae, such as Protochlamydia amoebophila (92), Parachlamydia acanthamoebae (93), and Neochlamydia hartmanellae (94). Coculture with amoebae also can identify new Chlamydia species from environmental samples, such as Criblamydia sequanensis, which was identified from a water sample (95). A recent study found that the chlamydial symbiont P. amoebophila can protect its Acanthamoeba castellanii host from L. pneumophila infection, indicating the interactions between amoebae and chlamydia may be more complicated than we expect (16). Since many chlamydiae can survive within amoebae, it is possible that amoebae can serve as shelters and environmental reservoirs for the transmission of chlamydiae and play a significant role in their lifecycles and ecology (96).
Mutualistic interactions.
Some ARBs established long-term relationships with amoebae as endosymbionts. Endosymbionts can stably reside within amoeba cells and survive encystations of the amoebae. For instance, 25% of natural Acanthamoeba isolates harbor obligate intracellular bacteria (12, 16, 97). However, it is challenging to disentangle the nature of their interactions, as most of the endosymbionts are unculturable, and they cannot grow outside their hosts. Therefore, most studies rely on nonculturable techniques, such as 16S rRNA gene sequencing or fluorescence in situ hybridization (FISH). Empirical studies on the cost and benefit of their interactions are needed.
To date, one of the best study systems to address the above question is the symbiosis of the social amoeba D. discoideum. D. discoideum is a soil-dwelling amoeba that is well known for its social life, consisting of a unicellular amoeboid stage and a multicellular aggregative stage in which about 20% of cells ultimately die to form a stalk to help spore dispersal (98). It has been used as a model species in cell biology and social evolution and is also one of the National Institutes of Health’s 13 model organisms. Recent studies found that some wild amoeba clones stably associate with different bacterial partners and use them as food and weapons (21, 26, 27). These clones are called farmers because they can seed and harvest their crops in new environments (21, 27). Two clades of inedible Burkholderia bacteria have been found to induce farming, causing the amoeba host to carry both them and edible crop bacteria (99–101). One of the clades shows considerable genome reduction (100). These Burkholderia organisms are facultative endosymbionts: they can live on their own and impose some cost on the host (21, 27, 102, 103). These Burkholderia symbionts also can preferentially find and choose their amoeba partners (18). Further studies find that amoeba hosts have evolved mechanisms for host-specific acquired symbionts and show evidence of partner adaptation between hosts and their symbionts (19, 102).
WHAT WE CAN LEARN FROM AMOEBA-BACTERIUM INTERACTIONS
Human pathogens and diseases.
The long history of interactions between amoebae and bacteria can select bacterial virulence traits that lead to the intracellular adaptation lifestyle (9). Therefore, amoebae can act as an environmental reservoir for pathogens and enable some ARBs to acquire the ability to infect mammals, including humans. Increasing studies use amoebae as host models to study human pathogens, and most studies used Acanthamoeba and Dictyostelium, both of which belong to the group Amoebozoa.
Using amoebae as host models for human pathogens can provide several insights. First, we can discover new virulence factors in amoebae using a simple plaque assay. For instance, the virulence of Pseudomonas aeruginosa was tested using amoebae (104). Different P. aeruginosa mutants were mixed with amoebae and then were plated on solid agar. If the strains were virulent, small amoebal lysis plaques would form. Several virulence factors, such as the quorum-sensing-mediated virulence factor lasR and the cytotoxin exoU, were discovered in P. aeruginosa (105). Second, we can isolate new bacterial species and investigate their pathogenicity in amoebae. Using amoebal coculture or amoebal enrichment, we can discover and isolate new ARBs directly from environmental samples, which can be used for subsequent pathogenic analysis (106). Finally, we can use amoebae to study host susceptibility and resistance to pathogenic infection. For instance, D. discoideum is an ideal model for such purposes because it has a fully sequenced haploid genome and is easy to manipulate genetically. Many D. discoideum genes, such as racH (intercellular spreading of mycobacteria), corA (intracellular growth), atg9 (macroautophagy), phg1 (intracellular killing), and kil1 (intracellular killing), have been reported to be essential for its interaction with bacterial pathogens (106).
HGT.
Horizontal gene transfer (HGT) is the transfer of genetic material between organisms other than reproduction, which is suggested to be a significant driver of genome evolution (107). Amoeba-bacterium interactions provide us an ideal chance to investigate interkingdom horizontal gene transfer, as both partners have gained genes from each other (78, 108, 109). There is plenty of evidence that amoeba-resisting bacteria acquired genes from their amoeba hosts (108). For instance, L. pneumophila can infect both amoebae and human macrophages, mediated by Legionella effectors proteins that play an essential role (76). Among them, L. pneumophila acquired one effector, legS2, from amoebae (110). This protein is related to the type IV Icm/Dot secretion system and the mitochondria, which helps L. pneumophila exploit its host (110). In another case, it has been proposed that Legionella longbeachae also acquired dhcR7 and dwf, the 7-dehydrocholesterol reductase, from its amoeba hosts (109, 111).
Amoebae can also integrate bacterial genes into their genome through horizontal gene transfer. In the social amoeba D. discoideum, 18 candidate horizontal gene transfers have been identified in its genome (112). These transferred domains have different forms, replacing or adding new functions to D. discoideum’s genome (112). In another study, the authors found that Acanthamoeba castellanii and Naegleria gruberi contained more horizontally acquired bacterial genes than Entamoeba histolytica and D. discoideum (113). These results suggest that horizontal gene transfer is an important factor shaping the genome of amoebae.
In addition, amoebae can also serve as an environmental niche that allows horizontal gene transfer between intracellular bacteria. For example, a study shows that Bartonella rattaustraliani and Rhizobium radiobacter can conjugate together within Acanthamoeba polyphaga, which provides direct evidence of genetic transfer (114). Another study using a bioinformatics approach finds eight mycobacterial open reading frames (ORFs) that are likely acquired from ARBs such as Proteobacteria and Firmicutes (115). These results suggest that amoebae can promote horizontal gene transfer and contribute to the creation of emerging pathogens.
Health risk in drinking water systems.
Amoebae are widespread in water and soil, but they have also been found in human-made water systems, such as hospital water networks, swimming pools, cooling towers, and drinking water systems (10, 82, 116, 117). Some amoebae are pathogenic and even lethal to humans (118, 119). The discovery of amoebae in drinking water systems is of concern. For example, four pathogenic Acanthamoeba species (Acanthamoeba culbertsoni, Acanthamoeba polyphaga, Acanthamoeba castellanii, and Acanthamoeba rhysodes) can infect humans and lead to keratitis and granulomatous amoebic encephalitis (GAE) (120), while Naegleria fowleri can infect human brains and causes a fatality rate of 97% in the United States (119).
In addition to pathogenic amoebae, nonpathogenic amoebae can also pose potential health risks because of their associations with pathogenic bacteria such as L. pneumophila (14, 74). In addition, amoebae are found to be resistant to disinfection in drinking water processes. For instance, Acanthamoeba cysts can resist exposure to up to 100 mg liter−1 chlorine for 10 min, which means our current drinking water disinfection (1 to 2 mg liter −1) will have a limited effect on these amoebae (121). Therefore, amoebae can transport and even protect a range of waterborne pathogens. Future studies should investigate the diversity of bacterial pathogens carried by amoebae.
A simple model system for symbiosis and coevolution.
Bacterial symbionts have essential effects on the fitness of eukaryotes, ranging from parasitism to mutualism (122). To better understand their interactions, we need simple model systems where the impact of different partners can be understood and manipulated (123). An ideal study system is the amoebae and their associated bacteria. Amoeba-bacterium interactions provide us a unique chance to study bacterium-eukaryote interactions. For instance, in social amoeba symbiosis, D. discoideum amoebae carry a minimicrobiome consisting of several bacteria species (21, 26, 27, 99). These carried bacteria partners can provide novel traits to their hosts, such as protofarming and defense (21, 26, 27). They also can have different fitness consequences for their hosts, making this a great system to investigate cooperation and conflict in both partners. This system is also suitable for studying how antagonistic interactions evolve to be more mutualistic (19).
Amoeba-bacterium interactions also give us a great chance to study the origin of the primary plastid. It is hypothesized that the origin of the chloroplast is because a nonphotosynthetic protist caught photosynthetic cyanobacteria as organelles. A freshwater amoeba, Paulinella chromatophora, gives us a unique opportunity to test this idea experimentally. P. chromatophora has taken two cyanobacteria as organelles, which are also called chromatophores. The chromatophores are considered organelles because their division is controlled by amoeba, and their genome is reduced by 2/3 (124–127). Although reduced, the chromatophore genome is still larger than most plastid genomes, indicating this endosymbiosis is still under way, which provides an excellent system to study the origin of organelles.
RESEARCH GAPS AND FUTURE DIRECTIONS
The hidden diversity.
Amoebae and their associated bacteria have proven to be useful study systems. Using traditional methodology and techniques, we have accumulated lots of knowledge over the past few decades. However, there are at least two levels of hidden diversity that need further investigation.
First, the diversity of amoebae in the environment is mostly unknown. Traditionally, the cultivation method is widely used for the identification of environmental amoebae. Axenic culture is one option, but it is laborious, and only a few species have been cultivated successfully (128–130). Another strategy is to use bacteria as a food source, as most amoebae are bacterivorous. In those studies, nonnutrient agar is seeded with microbes such as Escherichia coli, Klebsiella pneumoniae, and even Saccharomyces cerevisiae (24, 43, 117, 129, 131). Nevertheless, parameters such as food sources, grazing preference, and growth conditions can significantly affect the outcome of cultivation, which could create a bias for the real amoeba community.
A solution is to utilize next-generation sequencing techniques such as metagenomics. Several studies have performed this approach to study amoeba diversity in different environments (82, 117, 132, 133). Metagenomics is also suitable for large-scale studies, such as continental and global analyses, which are urgently needed at the moment, especially for soil amoebae (134, 135). However, there are also drawbacks, such as primer choice and copy number variations. Until now, no study has compared the efficacy of primers on amoebae. We also know very little about the variation of copy numbers (rRNA genes) in different amoebae, which is crucial to correct 18S rRNA gene sequencing bias. Such information is crucial to reveal the real community and diversity of amoebae, as the copy number can vary from one amoeba to another and from trophozoite to cyst. In conclusion, future studies should first address the issues of primer choice and copy number variations, and next-generation sequencing is the most promising technique to investigate the diversity of amoebae.
Second, the diversity of bacterial symbionts in amoebae is likely underestimated. Since all amoebae feed and interact with bacteria, we would expect to find bacterial symbionts in all major amoeba groups. However, to date, most studies focused on the group of Amoebozoa, more specifically on Acanthamoeba spp. and Dictyostelium spp. (Table 1). In Acanthamoeba, a wide range of bacteria have been isolated, including Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, Actinobacteria, and Chlamydiae (7, 12, 14). These bacterial symbionts occupy different host niches and have distinct lifestyles within Acanthamoeba. For instance, many studies have reported obligate symbionts in Acanthamoeba, and they can grow only within amoeba hosts (32, 89, 96).
However, we know very little about the bacterial symbionts in other groups of amoebae (Table 1). Several studies focused on the symbionts in Naegleria (14, 120, 136), probably because N. fowleri can cause a rare infection of the brain called primary amoebic meningoencephalitis (PAM). Other than that, only a few studies have reported bacterial symbionts in other groups of amoebae (Table 1), and there is a research gap that demands future studies. We expect that current studies underestimate the real diversity of bacterial symbionts in amoebae, because most current studies focus only on limited amoeba species. We need more studies on other amoeba groups, which can give fruitful information on the interactions between amoebae and bacteria. For instance, P. chromatophora, a freshwater amoeba that belonged to the Rhizaria group, recently (in evolutionary scales) caught a cyanobacterium and is transforming it into a photosynthetic organelle, which provides an excellent study system for the origin of the organelle (124, 126, 127). Given the long history of coevolution between amoebae and bacteria, future studies on other groups of amoebae may discover new types of interactions that cover the whole spectrum of symbiotic relationships.
More amoeba genomes needed.
The Genomes OnLine database currently has 375,306 species with sequenced genomes (137). Of these, 33,300 genomes are from eukaryotic organisms. However, most of these are from plants, animals, and fungi, and only 3% of them (1,151) belong to protists (137). That is probably why the nomenclature and taxonomy of protists are challenging, and their classification is being revised and continuously updated (3, 4, 138, 139).
For amoebae, to our knowledge, there are only 41 species that have sequenced genomes, and most of them come from Dictyostelium, Acanthamoeba, Naegleria, and Entamoeba (137). Amoeba genomes only account for 0.1% of the sequenced eukaryotes and 0.01% of all sequenced organisms, and many lineages of amoebae have no genomics information at all. Such a knowledge gap has dramatically hindered our understanding of the ecology and evolution of amoebae as well as their interactions with bacteria. Although barcoding techniques have begun to be applied in amoeba studies, they are not enough. These data may be sufficient for biogeographic investigation, but they cannot be used to test more specific hypotheses, such as how amoebae interact with their symbionts. With reduced sequencing cost, future studies should take this opportunity and sequence more amoeba genomes. More genomics data will help better understand the phylogeny of amoebae, the transfer of genes across genomes, and the ecology, evolution, and cell biology of amoeba-bacterium interactions.
Amoeba predation and bacterial microbiome.
Studies of macroorganisms such as animals have shown how important predators can be to the composition of ecological communities and ecosystems (140). Amoebae are important predators in some environments and, therefore, may have essential effects on the composition of bacterial communities. As we have come to realize the importance of bacterial microbiomes for the health of hosts, it is worth pursuing whether amoeba predators affect these relations. We briefly mention two possibilities among many.
Plant growth and health can be strongly impacted by the bacterial microbiome in the rhizosphere near the roots (141), and protists may have important effects (142). For example, the experimental addition of A. castellanii strongly affected the abundance and composition of Arabidopsis rhizosphere bacterial communities relative to controls with no protist predators (35). The presence of amoebae also increased Arabidopsis seedling growth. Part of this effect is that protists release excess nitrogen in the form of ammonium (143). More work is needed to investigate the role of changes in the bacterial community.
Amoebae might also be important in gut microbiomes. Various amoebae, especially Entamoeba, are commonly found in vertebrate guts (144). Some are pathogens, like E. histolytica, which can cause amoebic dysentery, and others, like Entamoeba dispar, are commensals that might contribute to normal healthy gut function. Given the importance of predators for maintaining prey diversity in many ecosystems (145) and the importance of bacterial microbiome diversity for human health, it seems reasonable to speculate that predators such as amoebae sometimes are beneficial (146). E. histolytica has been shown to have feeding preferences and, therefore, might affect gut bacterial diversity (44). In a study of gut microbiomes of West African populations, the presence or absence of Entamoeba (species undetermined) was associated with large differences in bacterial communities, such that community composition could predict Entamoeba presence with 79% accuracy (147). Individuals with Entamoeba had higher bacterial diversity in their guts and greater uniformity between individuals, perhaps indicating greater stability. If the absence of amoeba predators contributes to gut dysfunction, the reintroduction of them might help restore gut microbiome function (146).
CONCLUSIONS
It has been more than 200 years since the first report of an amoeba, and we have accumulated a large body of knowledge. Numerous studies have proven that amoebae are an essential component of the aquatic and terrestrial ecosystems and play a vital role in the transmission and evolution of bacterial pathogens. However, there are still research gaps, such as detecting hidden diversity and a lack of amoeba genomes. Future studies should benefit from the development of sequencing techniques and reduced sequencing costs to reveal the whole picture of amoeba biology and amoeba-bacterium interactions.
ACKNOWLEDGMENTS
We have no conflict of interest to declare.
L. Shu, Y. Shi, and C. Wang conceived the ideas and led the writing of the manuscript. S. Zhang contributed to the visualization. All authors contributed critically to the drafts and gave final approval for publication.
This material is based upon work supported by the National Natural Science Foundation of China (31970384, 41907021, and 21806044), the Fundamental Research Funds for the Central Universities (19lgpy162), the Guangdong Basic and Applied Basic Research Foundation (2019A1515011406), and the Hundred Talents Program through Sun Yat-Sen University (38000-18841205 and 99318-18841205).
Biographies

Yijing Shi completed her Ph.D. thesis at Shandong University and ETH (Eawag) and received her Ph.D. in environmental engineering. She had her postdoctoral training at the University of Alberta and was well trained in environmental microbiology. Since 2018, she has been an associate research fellow at South China Normal University. Her research interests focus on applying the knowledge of environmental microbiology and microbial ecology to a variety of environmental engineering applications.

David C. Queller earned a B.A. in history and philosophy of science from the University of Illinois and a Ph.D. in biological sciences from the University of Michigan, and he did postdoctoral work at the University of Sussex. He spent many years on the faculty at Rice University, moving to Washington University in 2011, where he is the Spencer T. Olin Professor of Biology. He has spent 40 years working in a variety of areas of evolutionary biology, especially the evolution of social interactions. For the last 20 years he has worked on the evolutionary biology of social amoebae, coleading a research group with Joan Strassmann. He is an elected Fellow of the American Association for the Advancement of Science and the American Academy of Arts and Sciences.

Cheng Wang received his Ph.D. in microbiology from Zhejiang University and completed his postdoctoral training in molecular microbial ecology at Max Planck Institute for Marine Microbiology as well. He is currently an associate research fellow at Sun Yat-Sen University, where he and his team focus on the development and implementation of novel solutions for omics data analysis. He has pursued a variety of research topics related to the microbiome and environmental pollution over the course of the last 10 years.

Longfei Shu earned his Ph.D. in environmental science at ETH Zürich. After that, he was a postdoctoral fellow in the Queller/Strassmann Research Group at Washington University in St. Louis. Since 2018, he has been an associate professor at Sun Yat-Sen University. He has spent over 10 years on ecology and environmental science. His main research interest is the ecology and evolution of amoebae, focusing on amoeba-microbe interactions (symbiosis) and amoeba-environment interactions (environmental pollution and climate changes).
REFERENCES
- 1.Levine ND, Corliss JO, Cox FE, Deroux G, Grain J, Honigberg BM, Leedale GF, Loeblich AR III, Lom J, Lynn D, Merinfeld EG, Page FC, Poljansky G, Sprague V, Vavra J, Wallace FG. 1980. A newly revised classification of the protozoa. J Protozool 27:37–58. doi: 10.1111/j.1550-7408.1980.tb04228.x. [DOI] [PubMed] [Google Scholar]
- 2.Pawlowski J. 2008. The twilight of Sarcodina: a molecular perspective on the polyphyletic origin of amoeboid protists. Protistology 5:281–302. [Google Scholar]
- 3.Adl SM, Bass D, Lane CE, Lukeš J, Schoch CL, Smirnov A, Agatha S, Berney C, Brown MW, Burki F, Cárdenas P, Čepička I, Chistyakova L, Del Campo J, Dunthorn M, Edvardsen B, Eglit Y, Guillou L, Hampl V, Heiss AA, Hoppenrath M, James TY, Karnkowska A, Karpov S, Kim E, Kolisko M, Kudryavtsev A, Lahr DJG, Lara E, Le Gall L, Lynn DH, Mann DG, Massana R, Mitchell EAD, Morrow C, Park JS, Pawlowski JW, Powell MJ, Richter DJ, Rueckert S, Shadwick L, Shimano S, Spiegel FW, Torruella G, Youssef N, Zlatogursky V, Zhang Q. 2019. Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol 66:4–119. doi: 10.1111/jeu.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adl SM, Leander BS, Simpson AGB, Archibald JM, Anderson OR, Bass D, Bowser SS, Brugerolle G, Farmer MA, Karpov S, Kolisko M, Lane CE, Lodge DJ, Mann DG, Meisterfeld R, Mendoza L, Moestrup O, Mozley-Standridge SE, Smirnov AV, Spiegel F. 2007. Diversity, nomenclature, and taxonomy of protists. Syst Biol 56:684–689. doi: 10.1080/10635150701494127. [DOI] [PubMed] [Google Scholar]
- 5.Kang S, Tice AK, Spiegel FW, Silberman JD, Panek T, Cepicka I, Kostka M, Kosakyan A, Alcantara DMC, Roger AJ, Shadwick LL, Smirnov A, Kudryavtsev A, Lahr DJG, Brown MW. 2017. Between a pod and a hard test: the deep evolution of amoebae. Mol Biol Evol 34:2258–2270. doi: 10.1093/molbev/msx162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jassey VEJ, Meyer C, Dupuy C, Bernard N, Mitchell EAD, Toussaint ML, Metian M, Chatelain AP, Gilbert D. 2013. To what extent do food preferences explain the trophic position of heterotrophic and mixotrophic microbial consumers in a sphagnum peatland? Microb Ecol 66:571–580. doi: 10.1007/s00248-013-0262-8. [DOI] [PubMed] [Google Scholar]
- 7.Samba-Louaka A, Delafont V, Rodier M-H, Cateau E, Héchard Y. 2019. Free-living amoebae and squatters in the wild: ecological and molecular features. FEMS Microbiol Rev 43:415–434. doi: 10.1093/femsre/fuz011. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez-Zaragoza S. 1994. Ecology of free-living amoebae. Crit Rev Microbiol 20:225–241. doi: 10.3109/10408419409114556. [DOI] [PubMed] [Google Scholar]
- 9.Strassmann JE, Shu L. 2017. Ancient bacteria-amoeba relationships and pathogenic animal bacteria. PLoS Biol 15:e2002460. doi: 10.1371/journal.pbio.2002460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas JM, Ashbolt NJ. 2011. Do free-living amoebae in treated drinking water systems present an emerging health risk? Environ Sci Technol 45:860–869. doi: 10.1021/es102876y. [DOI] [PubMed] [Google Scholar]
- 11.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horn M, Wagner M. 2004. Bacterial endosymbionts of free-living amoebae. J Eukaryot Microbiol 51:509–514. doi: 10.1111/j.1550-7408.2004.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 13.Greub G, Raoult D. 2004. Microorganisms resistant to free-living amoebae. Clin Microbiol Rev 17:413–433. doi: 10.1128/cmr.17.2.413-433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balczun C, Scheid PL. 2017. Free-living amoebae as hosts for and vectors of intracellular microorganisms with public health significance. Viruses 9:65. doi: 10.3390/v9040065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anderson OR. 2018. A half-century of research on free-living amoebae (1965–2017): review of biogeographic, ecological and physiological studies. Acta Protozool 57:1–28. doi: 10.4467/16890027AP.18.001.8395. [DOI] [Google Scholar]
- 16.König L, Wentrup C, Schulz F, Wascher F, Escola S, Swanson MS, Buchrieser C, Horn M. 2019. Symbiont-mediated defense against Legionella pneumophila in amoebae. mBio 10:e00333-19. doi: 10.1128/mBio.00333-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farinholt T, Dinh C, Kuspa A. 2019. Social amoebae establish a protective interface with their bacterial associates by lectin agglutination. Sci Adv 5:eaav4367. doi: 10.1126/sciadv.aav4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu L, Zhang B, Queller DC, Strassmann JE. 2018. Burkholderia bacteria use chemotaxis to find social amoeba Dictyostelium discoideum hosts. ISME J 12:1977–1993. doi: 10.1038/s41396-018-0147-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu L, Brock DA, Geist KS, Miller JW, Queller DC, Strassmann JE, DiSalvo S. 2018. Symbiont location, host fitness, and possible coadaptation in a symbiosis between social amoebae and bacteria. Elife 7:e42660. doi: 10.7554/eLife.42660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Zhuchenko O, Kuspa A, Soldati T. 2016. Social amoebae trap and kill bacteria by casting DNA nets. Nat Commun 7:10938. doi: 10.1038/ncomms10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brock DA, Douglas TE, Queller DC, Strassmann JE. 2011. Primitive agriculture in a social amoeba. Nature 469:393–396. doi: 10.1038/nature09668. [DOI] [PubMed] [Google Scholar]
- 22.Gerstenmaier L, Pilla R, Herrmann L, Herrmann H, Prado M, Villafano GJ, Kolonko M, Reimer R, Soldati T, King JS, Hagedorn M. 2015. The autophagic machinery ensures nonlytic transmission of mycobacteria. Proc Natl Acad Sci U S A 112:E687–E692. doi: 10.1073/pnas.1423318112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hagedorn M, Rohde KH, Russell DG, Soldati T. 2009. Infection by tubercular mycobacteria is spread by nonlytic ejection from their amoeba hosts. Science 323:1729–1733. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Delafont V, Bouchon D, Héchard Y, Moulin L. 2016. Environmental factors shaping cultured free-living amoebae and their associated bacterial community within drinking water network. Water Res 100:382–392. doi: 10.1016/j.watres.2016.05.044. [DOI] [PubMed] [Google Scholar]
- 25.Cosson P, Soldati T. 2008. Eat, kill or die: when amoeba meets bacteria. Curr Opin Microbiol 11:271–276. doi: 10.1016/j.mib.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 26.Stallforth P, Brock DA, Cantley AM, Tian XJ, Queller DC, Strassmann JE, Clardy J. 2013. A bacterial symbiont is converted from an inedible producer of beneficial molecules into food by a single mutation in the gacA gene. Proc Natl Acad Sci U S A 110:14528–14533. doi: 10.1073/pnas.1308199110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brock DA, Read S, Bozhchenko A, Queller DC, Strassmann JE. 2013. Social amoeba farmers carry defensive symbionts to protect and privatize their crops. Nat Commun 4:2385. doi: 10.1038/ncomms3385. [DOI] [PubMed] [Google Scholar]
- 28.Lima WC, Leuba F, Soldati T, Cosson P. 2012. Mucolipin controls lysosome exocytosis in Dictyostelium. J Cell Sci 125:2315–2322. doi: 10.1242/jcs.100362. [DOI] [PubMed] [Google Scholar]
- 29.Ray K, Marteyn B, Sansonetti PJ, Tang CM. 2009. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Rev Microbiol 7:333–340. doi: 10.1038/nrmicro2112. [DOI] [PubMed] [Google Scholar]
- 30.Boscaro V, Kolisko M, Felletti M, Vannini C, Lynn DH, Keeling PJ. 2017. Parallel genome reduction in symbionts descended from closely related free-living bacteria. Nat Ecol Evol 1:1160–1167. doi: 10.1038/s41559-017-0237-0. [DOI] [PubMed] [Google Scholar]
- 31.McCutcheon JP, Moran NA. 2011. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol 10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz-Esser S, Toenshoff ER, Haider S, Heinz E, Hoenninger VM, Wagner M, Horn M. 2008. Diversity of bacterial endosymbionts of environmental Acanthamoeba isolates. Appl Environ Microbiol 74:5822–5831. doi: 10.1128/AEM.01093-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cosson P, Lima WC. 2014. Intracellular killing of bacteria: is Dictyostelium a model macrophage or an alien? Cell Microbiol 16:816–823. doi: 10.1111/cmi.12291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bozzaro S, Eichinger L. 2011. The professional phagocyte Dictyostelium discoideum as a model host for bacterial pathogens. Curr Drug Targets 12:942–954. doi: 10.2174/138945011795677782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg K, Bertaux J, Krome K, Hartmann A, Scheu S, Bonkowski M. 2009. Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J 3:675–684. doi: 10.1038/ismej.2009.11. [DOI] [PubMed] [Google Scholar]
- 36.Matz C, Kjelleberg S. 2005. Off the hook−how bacteria survive protozoan grazing. Trends Microbiol 13:302–307. doi: 10.1016/j.tim.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Adiba S, Nizak C, van Baalen M, Denamur E, Depaulis F. 2010. From grazing resistance to pathogenesis: the coincidental evolution of virulence factors. PLoS One 5:e11882. doi: 10.1371/journal.pone.0011882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brock DA, Haselkorn TS, Garcia JR, Bashir U, Douglas TE, Galloway J, Brodie F, Queller DC, Strassmann JE. 2018. Diversity of free-living environmental bacteria and their interactions with a bactivorous amoeba. Front Cell Infect Microbiol 8:411. doi: 10.3389/fcimb.2018.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paquet VE, Charette SJ. 2016. Amoeba-resisting bacteria found in multilamellar bodies secreted by Dictyostelium discoideum: social amoebae can also package bacteria. FEMS Microbiol Ecol 92:fiw025. doi: 10.1093/femsec/fiw025. [DOI] [PubMed] [Google Scholar]
- 40.Anscombe FJ, Singh BN. 1948. Limitation of bacteria by micro-predators in soil. Nature 161:140. doi: 10.1038/161140a0. [DOI] [PubMed] [Google Scholar]
- 41.Eisenberg RM, Hurd LE, Ketcham RB. 1989. The cellular slime mold guild and its bacterial prey: growth rate variation at the inter- and intraspecific levels. Oecologia 79:458–462. doi: 10.1007/BF00378661. [DOI] [PubMed] [Google Scholar]
- 42.Khan NA, Siddiqui R. 2014. Predator vs aliens: bacteria interactions with Acanthamoeba. Parasitology 141:869–874. doi: 10.1017/S003118201300231X. [DOI] [PubMed] [Google Scholar]
- 43.Weekers PHH, Bodelier PLE, Wijen JPH, Vogels GD. 1993. Effects of grazing by the free-living soil amoebae Acanthamoeba castellanii, Acanthamoeba polyphaga, and Hartmannella vermiformis on various bacteria. Appl Environ Microbiol 59:2317–2319. doi: 10.1128/AEM.59.7.2317-2319.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iyer LR, Verma AK, Paul J, Bhattacharya A. 2019. Phagocytosis of gut bacteria by Entamoeba histolytica. Front Cell Infect Microbiol 9:34. doi: 10.3389/fcimb.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubin M, Miller AD, Katoh-Kurasawa M, Dinh C, Kuspa A, Shaulsky G. 2019. Cooperative predation in the social amoebae Dictyostelium discoideum. PLoS One 14:e0209438. doi: 10.1371/journal.pone.0209438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nasser W, Santhanam B, Miranda ER, Parikh A, Juneja K, Rot G, Dinh C, Chen R, Zupan B, Shaulsky G, Kuspa A. 2013. Bacterial discrimination by dictyostelid amoebae reveals the complexity of ancient interspecies interactions. Curr Biol 23:862–872. doi: 10.1016/j.cub.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lamrabet O, Melotti A, Burdet F, Hanna N, Perrin J, Nitschke J, Pagni M, Hilbi H, Soldati T, Cosson P. 2020. Transcriptional responses of Dictyostelium discoideum exposed to different classes of bacteria. Front Microbiol 11:410. doi: 10.3389/fmicb.2020.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, Lardy B, Klein G, Sansonetti P, Charette SJ, Cosson P. 2006. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol 8:139–148. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- 49.Rashidi G, Ostrowski EA. 2019. Phagocyte chase behaviours: discrimination between Gram-negative and Gram-positive bacteria by amoebae. Biol Lett 15:20180607. doi: 10.1098/rsbl.2018.0607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen GK, Zhuchenko O, Kuspa A. 2007. Immune-like phagocyte activity in the social amoeba. Science 317:678–681. doi: 10.1126/science.1143991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lima WC, Balestrino D, Forestier C, Cosson P. 2014. Two distinct sensing pathways allow recognition of Klebsiella pneumoniae by Dictyostelium amoebae. Cell Microbiol 16:311–323. doi: 10.1111/cmi.12226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bodinier R, Leiba J, Sabra A, Jauslin TN, Lamrabet O, Guilhen C, Marchetti A, Iwade Y, Kawata T, Lima WC, Cosson P. 2020. LrrkA, a kinase with leucine-rich repeats, links folate sensing with Kil2 activity and intracellular killing. Cell Microbiol 22:e13129. doi: 10.1111/cmi.13129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leiba J, Sabra A, Bodinier R, Marchetti A, Lima WC, Melotti A, Perrin J, Burdet F, Pagni M, Soldati T, Lelong E, Cosson P. 2017. Vps13F links bacterial recognition and intracellular killing in Dictyostelium. Cell Microbiol 19:e12722. doi: 10.1111/cmi.12722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuburich NA, Adhikari N, Hadwiger JA. 2016. Acanthamoeba and Dictyostelium use different foraging strategies. Protist 167:511–525. doi: 10.1016/j.protis.2016.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Russell DG, VanderVen BC, Glennie S, Mwandumba H, Heyderman RS. 2009. The macrophage marches on its phagosome: dynamic assays of phagosome function. Nat Rev Immunol 9:594–600. doi: 10.1038/nri2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marshansky V, Futai M. 2008. The V-type H+-ATPase in vesicular trafficking: targeting, regulation and function. Curr Opin Cell Biol 20:415–426. doi: 10.1016/j.ceb.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Journet A, Chapel A, Jehan S, Adessi C, Freeze H, Klein G, Garin J. 1999. Characterization of Dictyostelium discoideum cathepsin D. J Cell Sci 112:3833–3843. [DOI] [PubMed] [Google Scholar]
- 58.Muller I, Subert N, Otto H, Herbst R, Ruhling H, Maniak M, Leippe M. 2005. A Dictyostelium mutant with reduced lysozyme levels compensates by increased phagocytic activity. J Biol Chem 280:10435–10443. doi: 10.1074/jbc.M411445200. [DOI] [PubMed] [Google Scholar]
- 59.Hao XL, Luthje F, Ronn R, German NA, Li XJ, Huang FY, Kisaka J, Huffman D, Alwathnani HA, Zhu YG, Rensing C. 2016. A role for copper in protozoan grazing−two billion years selecting for bacterial copper resistance. Mol Microbiol 102:628–641. doi: 10.1111/mmi.13483. [DOI] [PubMed] [Google Scholar]
- 60.German N, Doyscher D, Rensing C. 2013. Bacterial killing in macrophages and amoeba: do they all use a brass dagger? Future Microbiol 8:1257–1264. doi: 10.2217/fmb.13.100. [DOI] [PubMed] [Google Scholar]
- 61.Lardy B, Bof M, Aubry L, Paclet MH, Morel F, Satre M, Klein G. 2005. NADPH oxidase homologs are required for normal cell differentiation and morphogenesis in Dictyostelium discoideum. Biochim Biophys Acta 1744:199–212. doi: 10.1016/j.bbamcr.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 62.Dunn JD, Bosmani C, Barisch C, Raykov L, Lefrancois LH, Cardenal-Munoz E, Lopez-Jimenez AT, Soldati T. 2017. Eat prey, live: Dictyostelium discoideum as a model for cell-autonomous defenses. Front Immunol 8:1906. doi: 10.3389/fimmu.2017.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris E, Wang N, Wu Wl WL, Weatherford A, De Lozanne A, Cardelli J. 2002. Dictyostelium LvsB mutants model the lysosomal defects associated with Chediak-Higashi syndrome. Mol Biol Cell 13:656–669. doi: 10.1091/mbc.01-09-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peracino B, Wagner C, Balest A, Balbo A, Pergolizzi B, Noegel AA, Steinert M, Bozzaro S. 2006. Function and mechanism of action of Dictyostelium Nramp1 (Slc11a1) in bacterial infection. Traffic 7:22–38. doi: 10.1111/j.1600-0854.2005.00356.x. [DOI] [PubMed] [Google Scholar]
- 65.Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, May RC, Soldati T, Machesky LM, Insall RH. 2011. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J Cell Biol 193:831–839. doi: 10.1083/jcb.201009119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winterbourn CC, Kettle AJ. 2013. Redox reactions and microbial killing in the neutrophil phagosome. Antioxid Redox Signal 18:642–660. doi: 10.1089/ars.2012.4827. [DOI] [PubMed] [Google Scholar]
- 67.Lelong E, Marchetti A, Gueho A, Lima WC, Sattler N, Molmeret M, Hagedorn M, Soldati T, Cosson P. 2011. Role of magnesium and a phagosomal P-type ATPase in intracellular bacterial killing. Cell Microbiol 13:246–258. doi: 10.1111/j.1462-5822.2010.01532.x. [DOI] [PubMed] [Google Scholar]
- 68.Cornillon S, Dubois A, Bruckert F, Lefkir Y, Marchetti A, Benghezal M, De Lozanne A, Letourneur F, Cosson P. 2002. Two members of the beige/CHS (BEACH) family are involved at different stages in the organization of the endocytic pathway in Dictyostelium. J Cell Sci 115:737–744. [DOI] [PubMed] [Google Scholar]
- 69.Price CTD, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. 2011. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 70.Best AM, Abu Kwaik Y. 2019. Evasion of phagotrophic predation by protist hosts and innate immunity of metazoan hosts by Legionella pneumophila. Cell Microbiol 21:e12971. doi: 10.1111/cmi.12971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagele S, Kohler R, Merkert H, Schleicher M, Hacker J, Steinert M. 2000. Dictyostelium discoideum: a new host model system for intracellular pathogens of the genus Legionella. Cell Microbiol 2:165–171. doi: 10.1046/j.1462-5822.2000.00044.x. [DOI] [PubMed] [Google Scholar]
- 72.Solomon JM, Rupper A, Cardelli JA, Isberg RR. 2000. Intracellular growth of Legionella pneumophila in Dictyostelium discoideum, a system for genetic analysis of host-pathogen interactions. Infect Immun 68:2939–2947. doi: 10.1128/iai.68.5.2939-2947.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bozue JA, Johnson W. 1996. Interaction of Legionella pneumophila with Acanthamoeba castellanii: uptake by coiling phagocytosis and inhibition of phagosome lysosome fusion. Infect Immun 64:668–673. doi: 10.1128/IAI.64.2.668-673.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mengue L, Regnacq M, Aucher W, Portier E, Hechard Y, Samba-Louaka A. 2016. Legionella pneumophila prevents proliferation of its natural host Acanthamoeba castellanii. Sci Rep 6:36448. doi: 10.1038/srep36448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoffmann C, Harrison CF, Hilbi H. 2014. The natural alternative: protozoa as cellular models for Legionella infection. Cell Microbiol 16:15–26. doi: 10.1111/cmi.12235. [DOI] [PubMed] [Google Scholar]
- 76.Best A, Abu Kwaik Y. 2018. Evolution of the arsenal of Legionella pneumophila effectors to modulate protist hosts. mBio 9:e01313-18. doi: 10.1128/mBio.01313-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Price CTD, Al-Quadan T, Santic M, Jones SC, Abu Kwaik Y. 2010. Exploitation of conserved eukaryotic host cell farnesylation machinery by an F-box effector of Legionella pneumophila. J Exp Med 207:1713–1725. doi: 10.1084/jem.20100771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Price CT, Al-Khodor S, Al-Quadan T, Santic M, Habyarimana F, Kalia A, Abu Kwaik Y. 2009. Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog 5:e1000704. doi: 10.1371/journal.ppat.1000704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Al-Quadan T, Abu Kwaik Y. 2011. Molecular characterization of exploitation of the polyubiquitination and farnesylation machineries of Dictyostelium discoideum by the AnkB F-box effector of Legionella pneumophila. Front Microbiol 2:23. doi: 10.3389/fmicb.2011.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Price C, Jones S, Mihelcic M, Santic M, Abu Kwaik Y. 2020. Paradoxical pro-inflammatory responses by human macrophages to an amoebae host-adapted Legionella effector. Cell Host Microbe 27:571–584. doi: 10.1016/j.chom.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cervero-Arago S, Sommer R, Araujo RM. 2014. Effect of UV irradiation (253.7 nm) on free Legionella and Legionella associated with its amoebae hosts. Water Res 67:299–309. doi: 10.1016/j.watres.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 82.Delafont V, Mougari F, Cambau E, Joyeux M, Bouchon D, Hechard Y, Moulin L. 2014. First evidence of amoebae-mycobacteria association in drinking water network. Environ Sci Technol 48:11872–11882. doi: 10.1021/es5036255. [DOI] [PubMed] [Google Scholar]
- 83.Eddyani M, De Jonckheere JF, Durnez L, Suykerbuyk P, Leirs H, Portaels F. 2008. Occurrence of free-living amoebae in communities of low and high endemicity for buruli ulcer in southern Benin. Appl Environ Microbiol 74:6547–6553. doi: 10.1128/AEM.01066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thomas V, Loret JF, Jousset M, Greub G. 2008. Biodiversity of amoebae and amoebae-resisting bacteria in a drinking water treatment plant. Environ Microbiol 10:2728–2745. doi: 10.1111/j.1462-2920.2008.01693.x. [DOI] [PubMed] [Google Scholar]
- 85.Ovrutsky AR, Chan ED, Kartalija M, Bai XY, Jackson M, Gibbs S, Falkinham JO, Iseman MD, Reynolds PR, McDonnell G, Thomas V. 2013. Cooccurrence of free-living amoebae and nontuberculous mycobacteria in hospital water networks, and preferential growth of Mycobacterium avium in Acanthamoeba lenticulata. Appl Environ Microbiol 79:3185–3192. doi: 10.1128/AEM.03823-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pagnier I, Raoult D, La Scola B. 2008. Isolation and identification of amoeba-resisting bacteria from water in human environment by using an Acanthamoeba polyphaga co-culture procedure. Environ Microbiol 10:1135–1144. doi: 10.1111/j.1462-2920.2007.01530.x. [DOI] [PubMed] [Google Scholar]
- 87.Butler RE, Smith AA, Mendum TA, Chandran A, Wu HH, Lefrancois L, Chambers M, Soldati T, Stewart GR. 2020. Mycobacterium bovis uses the ESX-1 Type VII secretion system to escape predation by the soil-dwelling amoeba Dictyostelium discoideum. ISME J 14:919–930. doi: 10.1038/s41396-019-0572-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lamrabet O, Drancourt M. 2013. Mycobacterium gilvum illustrates size-correlated relationships between mycobacteria and Acanthamoeba polyphaga. Appl Environ Microbiol 79:1606–1611. doi: 10.1128/AEM.03765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Elwell C, Mirrashidi K, Engel J. 2016. Chlamydia cell biology and pathogenesis. Nat Rev Microbiol 14:385–400. doi: 10.1038/nrmicro.2016.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Connell CM, Ferone ME. 2009. Genital Chlamydia trachomatis infections. Clin Microbiol Infect 15:4–10. doi: 10.1111/j.1469-0691.2008.02647.x. [DOI] [PubMed] [Google Scholar]
- 91.Kalman S, Mitchell W, Marathe R, Lammel C, Fan L, Hyman RW, Olinger L, Grimwood L, Davis RW, Stephens RS. 1999. Comparative genomes of Chlamydia pneumoniae and C-trachomatis. Nat Genet 21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 92.Collingro A, Toenshoff ER, Taylor MW, Fritsche TR, Wagner M, Horn M. 2005. “Candidatus Protochlamydia amoebophila”, an endosymbiont of Acanthamoeba spp. Int J Syst Evol Microbiol 55:1863–1866. doi: 10.1099/ijs.0.63572-0. [DOI] [PubMed] [Google Scholar]
- 93.Everett KD, Bush RM, Andersen AA. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol 49(Pt 2):415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 94.Horn M, Wagner M, Muller KD, Schmid EN, Fritsche TR, Schleifer KH, Michel R. 2000. Neochlamydia hartmannellae gen. nov., sp nov (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231–1239. doi: 10.1099/00221287-146-5-1231. [DOI] [PubMed] [Google Scholar]
- 95.Thomas V, Casson N, Greub G. 2006. Criblamydia sequanensis, a new intracellular Chlamydiales isolated from Seine river water using amoebal co-culture. Environ Microbiol 8:2125–2135. doi: 10.1111/j.1462-2920.2006.01094.x. [DOI] [PubMed] [Google Scholar]
- 96.Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol 62:113–131. doi: 10.1146/annurev.micro.62.081307.162818. [DOI] [PubMed] [Google Scholar]
- 97.Schulz F, Horn M. 2015. Intranuclear bacteria: inside the cellular control center of eukaryotes. Trends Cell Biol 25:339–346. doi: 10.1016/j.tcb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 98.Kessin RH. 2001. Dictyostelium: evolution, cell biology, and the development of multicellularity. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 99.DiSalvo S, Haselkorn TS, Bashir U, Jimenez D, Brock DA, Queller DC, Strassmann JE. 2015. Burkholderia bacteria infectiously induce the proto-farming symbiosis of Dictyostelium amoebae and food bacteria. Proc Natl Acad Sci U S A 112:E5029–E5037. doi: 10.1073/pnas.1511878112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brock DA, Noh S, Hubert ANM, Haselkorn TS, DiSalvo S, Suess MK, Bradley AS, Tavakoli-Nezhad M, Geist KS, Queller DC, Strassmann JE. 2020. Endosymbiotic adaptations in three new bacterial species associated with Dictyostelium discoideum: Paraburkholderia agricolaris sp. nov., Paraburkholderia hayleyella sp. nov., and Paraburkholderia bonniea sp. nov. PeerJ 8:e9151. doi: 10.7717/peerj.9151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Garcia JR, Larsen TJ, Queller DC, Strassmann JE. 2019. Fitness costs and benefits vary for two facultative Burkholderia symbionts of the social amoeba, Dictyostelium discoideum. Ecol Evol 9:9878–9890. doi: 10.1002/ece3.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Haselkorn TS, DiSalvo S, Miller JW, Bashir U, Brock DA, Queller DC, Strassmann JE. 2019. The specificity of Burkholderia symbionts in the social amoeba farming symbiosis: prevalence, species, genetic and phenotypic diversity. Mol Ecol 28:847–862. doi: 10.1111/mec.14982. [DOI] [PubMed] [Google Scholar]
- 103.Shu L, Qian X, Brock DA, Geist KS, Queller DC, Strassmann JE. 20 October 2020. Loss and resiliency of social amoeba symbiosis under simulated warming. Ecol Evol doi: 10.1002/ece3.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pukatzki S, Kessin RH, Mekalanos JJ. 2002. The human pathogen Pseudomonas aeruginosa utilizes conserved virulence pathways to infect the social amoeba Dictyostelium discoideum. Proc Natl Acad Sci U S A 99:3159–3164. doi: 10.1073/pnas.052704399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dondero F, Jonsson H, Rebelo M, Pesce G, Berti E, Pons G, Viarengo A. 2006. Cellular responses to environmental contaminants in amoebic cells of the slime mould Dictyostelium discoideum. Comp Biochem Physiol C Toxicol Pharmacol 143:150–157. doi: 10.1016/j.cbpc.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 106.Tosetti N, Croxatto A, Greub G. 2014. Amoebae as a tool to isolate new bacterial species, to discover new virulence factors and to study the host-pathogen interactions. Microb Pathog 77:125–130. doi: 10.1016/j.micpath.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 107.Husnik F, McCutcheon JP. 2018. Functional horizontal gene transfer from bacteria to eukaryotes. Nat Rev Microbiol 16:67–79. doi: 10.1038/nrmicro.2017.137. [DOI] [PubMed] [Google Scholar]
- 108.Bertelli C, Greub G. 2012. Lateral gene exchanges shape the genomes of amoeba-resisting microorganisms. Front Cell Infect Microbiol 2:110. doi: 10.3389/fcimb.2012.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moliner C, Fournier PE, Raoult D. 2010. Genome analysis of microorganisms living in amoebae reveals a melting pot of evolution. FEMS Microbiol Rev 34:281–294. doi: 10.1111/j.1574-6976.2010.00209.x. [DOI] [PubMed] [Google Scholar]
- 110.Degtyar E, Zusman T, Ehrlich M, Segal G. 2009. A Legionella effector acquired from protozoa is involved in sphingolipids metabolism and is targeted to the host cell mitochondria. Cell Microbiol 11:1219–1235. doi: 10.1111/j.1462-5822.2009.01328.x. [DOI] [PubMed] [Google Scholar]
- 111.Thomas V, Greub G. 2010. Amoeba/amoebal symbiont genetic transfers: lessons from giant virus neighbours. Intervirology 53:254–267. doi: 10.1159/000312910. [DOI] [PubMed] [Google Scholar]
- 112.Eichinger L, Pachebat JA, Glöckner G, Rajandream M-A, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, van Driessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, Hauser H, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Clarke M, Lohan AJ, Liu B, Lagkouvardos I, Roy S, Zafar N, Bertelli C, Schilde C, Kianianmomeni A, Burglin TR, Frech C, Turcotte B, Kopec KO, Synnott JM, Choo C, Paponov I, Finkler A, Tan CSH, Hutchins AP, Weinmeier T, Rattei T, Chu JS, Gimenez G, Irimia M, Rigden DJ, Fitzpatrick DA, Lorenzo-Morales J, Bateman A, Chiu CH, Tang P, Hegemann P, Fromm H, Raoult D, Greub G, Miranda-Saavedra D, Chen N, Nash P, Ginger ML, Horn M, Schaap P, Caler L, Loftus BJ. 2013. Genome of Acanthamoeba castellanii highlights extensive lateral gene transfer and early evolution of tyrosine kinase signaling. Genome Biol 14:R11. doi: 10.1186/gb-2013-14-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Saisongkorh W, Robert C, La Scola B, Raoult D, Rolain JM. 2010. Evidence of transfer by conjugation of type IV secretion system genes between Bartonella species and Rhizobium radiobacter in amoeba. PLoS One 5:e12666. doi: 10.1371/journal.pone.0012666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lamrabet O, Merhej V, Pontarotti P, Raoult D, Drancourt M. 2012. The genealogic tree of mycobacteria reveals a long-standing sympatric life into free-living protozoa. PLoS One 7:e34754. doi: 10.1371/journal.pone.0034754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Thomas V, McDonnell G, Denyer SP, Maillard JY. 2010. Free-living amoebae and their intracellular pathogenic microorganisms: risks for water quality. FEMS Microbiol Rev 34:231–259. doi: 10.1111/j.1574-6976.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 117.Delafont V, Brouke A, Bouchon D, Moulin L, Hechard Y. 2013. Microbiome of free-living amoebae isolated from drinking water. Water Res 47:6958–6965. doi: 10.1016/j.watres.2013.07.047. [DOI] [PubMed] [Google Scholar]
- 118.Cooper AM, Aouthmany S, Shah K, Rega PP. 2019. Killer amoebas: primary amoebic meningoencephalitis in a changing climate. JAAPA 32:30–35. doi: 10.1097/01.JAA.0000558238.99250.4a. [DOI] [PubMed] [Google Scholar]
- 119.Bartrand TA, Causey JJ, Clancy JL. 2014. Naegleria fowleri: an emerging drinking water pathogen. J Am Water Works Assoc 106:418–432. doi: 10.5942/jawwa.2014.106.0140. [DOI] [Google Scholar]
- 120.Visvesvara GS, Moura H, Schuster FL. 2007. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol 50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 121.Dupuy M, Mazoua S, Berne F, Bodet C, Garrec N, Herbelin P, Menard-Szczebara F, Oberti S, Rodier MH, Soreau S, Wallet F, Hechard Y. 2011. Efficiency of water disinfectants against Legionella pneumophila and Acanthamoeba. Water Res 45:1087–1094. doi: 10.1016/j.watres.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 122.McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moran NA. 2006. Symbiosis. Curr Biol 16:R866–871. doi: 10.1016/j.cub.2006.09.019. [DOI] [PubMed] [Google Scholar]
- 124.Nowack ECM, Price DC, Bhattacharya D, Singer A, Melkonian M, Grossman AR. 2016. Gene transfers from diverse bacteria compensate for reductive genome evolution in the chromatophore of Paulinella chromatophora. Proc Natl Acad Sci U S A 113:12214–12219. doi: 10.1073/pnas.1608016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bhattacharya D, Price DC, Yoon HS, Yang EC, Poulton NJ, Andersen RA, Das SP. 2012. Single cell genome analysis supports a link between phagotrophy and primary plastid endosymbiosis. Sci Rep 2:356. doi: 10.1038/srep00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Singer A, Poschmann G, Muhlich C, Valadez-Cano C, Hansch S, Huren V, Rensing SA, Stuhler K, Nowack ECM. 2017. Massive protein import into the early-evolutionary-stage photosynthetic organelle of the amoeba Paulinella chromatophora. Curr Biol 27:2763–2773. doi: 10.1016/j.cub.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 127.Nakayama T, Ishida K. 2009. Another acquisition of a primary photosynthetic organelle is underway in Paulinella chromatophora. Curr Biol 19:R284–285. doi: 10.1016/j.cub.2009.02.043. [DOI] [PubMed] [Google Scholar]
- 128.Fields BS, Nerad TA, Sawyer TK, King CH, Barbaree JM, Martin WT, Morrill WE, Sanden GN. 1990. Characterization of an axenic strain of Hartmannella vermiformis obtained from an investigation of nosocomial legionellosis. J Protozool 37:581–583. doi: 10.1111/j.1550-7408.1990.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 129.Schuster FL. 2002. Cultivation of pathogenic and opportunistic free-living amoebas. Clin Microbiol Rev 15:342–354. doi: 10.1128/cmr.15.3.342-354.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schuster FL, Visvesvara GS. 1996. Axenic growth and drug sensitivity studies of Balamuthia mandrillaris, an agent of amebic meningoencephalitis in humans and other animals. J Clin Microbiol 34:385–388. doi: 10.1128/JCM.34.2.385-388.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Delafont V, Rodier MH, Maisonneuve E, Cateau E. 2018. Vermamoeba vermiformis: a free-living amoeba of interest. Microb Ecol 76:991–1001. doi: 10.1007/s00248-018-1199-8. [DOI] [PubMed] [Google Scholar]
- 132.Ren K, Xue Y, Rønn R, Liu L, Chen H, Rensing C, Yang J. 2018. Dynamics and determinants of amoeba community, occurrence and abundance in subtropical reservoirs and rivers. Water Res 146:177–186. doi: 10.1016/j.watres.2018.09.011. [DOI] [PubMed] [Google Scholar]
- 133.Delafont V, Perrin Y, Bouchon D, Moulin L, Hechard Y. 2019. Targeted metagenomics of microbial diversity in free-living amoebae and water samples. Methods Mol Biol 1921:421–428. doi: 10.1007/978-1-4939-9048-1_26. [DOI] [PubMed] [Google Scholar]
- 134.Geisen S, Mitchell EAD, Adl S, Bonkowski M, Dunthorn M, Ekelund F, Fernandez LD, Jousset A, Krashevska V, Singer D, Spiegel FW, Walochnik J, Lara E. 2018. Soil protists: a fertile frontier in soil biology research. FEMS Microbiol Rev 42:293–323. doi: 10.1093/femsre/fuy006. [DOI] [PubMed] [Google Scholar]
- 135.Geisen S, Mitchell EAD, Wilkinson DM, Adl S, Bonkowski M, Brown MW, Fiore-Donno AM, Heger TJ, Jassey VEJ, Krashevska V, Lahr DJG, Marcisz K, Mulot M, Payne R, Singer D, Anderson OR, Charman DJ, Ekelund F, Griffiths BS, Ronn R, Smirnov A, Bass D, Belbahri L, Berney C, Blandenier Q, Chatzinotas A, Clarholm M, Dunthorn M, Feest A, Fernandez LD, Foissner W, Fournier B, Gentekaki E, Hajek M, Helder J, Jousset A, Koller R, Kumar S, La Terza A, Lamentowicz M, Mazei Y, Santos SS, Seppey CVW, Spiegel FW, Walochnik J, Winding A, Lara E. 2017. Soil protistology rebooted: 30 fundamental questions to start with. Soil Biol Biochem 111:94–103. doi: 10.1016/j.soilbio.2017.04.001. [DOI] [Google Scholar]
- 136.Corsaro D, Muller KD, Michel R. 2013. Molecular characterization and ultrastructure of a new amoeba endoparasite belonging to the Stenotrophomonas maltophilia complex. Exp Parasitol 133:383–390. doi: 10.1016/j.exppara.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 137.Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Katta HY, Mojica A, Chen IMA, Kyrpides NC, Reddy TBK. 2019. Genomes OnLine database (GOLD) v.7: updates and new features. Nucleic Acids Res 47:D649–D659. doi: 10.1093/nar/gky977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EAD, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Shadwick L, Schoch CL, Smirnov A, Spiegel FW. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol 59:429–493. doi: 10.1111/j.1550-7408.2012.00644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Burki F, Roger AJ, Brown MW, Simpson AGB. 2020. The new tree of eukaryotes. Trends Ecol Evol 35:43–55. doi: 10.1016/j.tree.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 140.Terborgh J, Estes JA. 2013. Trophic cascades: predators, prey and the changing dynamics of nature. Island Press, Washington, DC. [Google Scholar]
- 141.Berendsen RL, Pieterse CMJ, Bakker PAHM. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 142.Gao Z, Karlsson I, Geisen S, Kowalchuk G, Jousset A. 2019. Protists: puppet masters of the rhizosphere microbiome. Trends Plant Sci 24:165–176. doi: 10.1016/j.tplants.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 143.Ekelund F, Ronn R. 1994. Notes on protozoa in agricultural soil with emphasis on heterotrophic flagellates and naked amoebae and their ecology. FEMS Microbiol Rev 15:321–353. doi: 10.1111/j.1574-6976.1994.tb00144.x. [DOI] [PubMed] [Google Scholar]
- 144.Parfrey LW, Walters WA, Knight R. 2011. Microbial eukaryotes in the human microbiome: ecology, evolution, and future directions. Front Microbiol 2:153. doi: 10.3389/fmicb.2011.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Vidal MC, Murphy SM. 2018. Bottom-up vs. top-down effects on terrestrial insect herbivores: a meta-analysis. Ecol Lett 21:138–150. doi: 10.1111/ele.12874. [DOI] [PubMed] [Google Scholar]
- 146.Mosca A, Leclerc M, Hugot JP. 2016. Gut microbiota diversity and human diseases: should we reintroduce key predators in our ecosystem? Front Microbiol 7:455. doi: 10.3389/fmicb.2016.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Morton ER, Lynch J, Froment A, Lafosse S, Heyer E, Przeworski M, Blekhman R, Segurel L. 2015. Variation in rural african gut microbiota is strongly correlated with colonization by entamoeba and subsistence. PLoS Genet 11:e1005658. doi: 10.1371/journal.pgen.1005658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Okubo T, Matsushita M, Nakamura S, Matsuo J, Nagai H, Yamaguchi H. 2018. Acanthamoeba S13WT relies on its bacterial endosymbiont to backpack human pathogenic bacteria and resist Legionella infection on solid media. Environ Microbiol Rep 10:344–354. doi: 10.1111/1758-2229.12645. [DOI] [PubMed] [Google Scholar]
- 149.Inglis RF, Asikhia O, Ryu E, Queller DC, Strassmann JE. 2018. Predator-by-environment interactions mediate bacterial competition in the Dictyostelium discoideum microbiome. Front Microbiol 9:781. doi: 10.3389/fmicb.2018.00781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Taylor-Mulneix DL, Bendor L, Linz B, Rivera I, Ryman VE, Dewan KK, Wagner SM, Wilson EF, Hilburger LJ, Cuff LE, West CM, Harvill ET. 2017. Bordetella bronchiseptica exploits the complex life cycle of Dictyostelium discoideum as an amplifying transmission vector. PLoS Biol 15:e2000420. doi: 10.1371/journal.pbio.2000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Schulz F, Lagkouvardos I, Wascher F, Aistleitner K, Kostanjsek R, Horn M. 2014. Life in an unusual intracellular niche: a bacterial symbiont infecting the nucleus of amoebae. ISME J 8:1634–1644. doi: 10.1038/ismej.2014.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Michel R, Müller K-D, Zöller L, Walochnik J, Hartmann M, Schmid E-N. 2005. Free-living amoebae serve as host for the Chlamydia-like Simkania negevensis. Acta Protozool 44:113–121. [Google Scholar]
- 153.Schulz F, Tyml T, Pizzetti I, Dykova I, Fazi S, Kostka M, Horn M. 2015. Marine amoebae with cytoplasmic and perinuclear symbionts deeply branching in the Gammaproteobacteria. Sci Rep 5:13381. doi: 10.1038/srep13381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Corsaro D, Müller K-D, Wingender J, Michel R. 2013. “Candidatus Mesochlamydia elodeae” (Chlamydiae: Parachlamydiaceae), a novel chlamydia parasite of free-living amoebae. Parasitol Res 112:829–838. doi: 10.1007/s00436-012-3213-2. [DOI] [PubMed] [Google Scholar]
- 155.Michel R, Müller K-D, Schmid E. 1995. Ehrlichia-like organismus (KSL1) observed as obligate intracellular parasites of Saccamoeba species. Endocytobiosis Cell Res 11:69–80. [Google Scholar]
- 156.Tsao HF, Scheikl U, Volland JM, Kohsler M, Bright M, Walochnik J, Horn M. 2017. “Candidatus Cochliophilus cryoturris” (Coxiellaceae), a symbiont of the testate amoeba Cochliopodium minus. Sci Rep 7:3394. doi: 10.1038/s41598-017-03642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gutierrez G, Chistyakova LV, Villalobo E, Kostygov AY, Frolov AO. 2017. Identification of Pelomyxa palustris endosymbionts. Protist 168:408–424. doi: 10.1016/j.protis.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 158.Park M, Yun ST, Kim MS, Chun J, Ahn TI. 2004. Phylogenetic characterization of Legionella-like endosymbiotic X-bacteria in Amoeba proteus: a proposal for “Candidatus Legionella jeonii” sp. nov. Environ Microbiol 6:1252–1263. doi: 10.1111/j.1462-2920.2004.00659.x. [DOI] [PubMed] [Google Scholar]
- 159.Broers CA, Meijers HH, Symens JC, Stumm CK, Vogels GD, Brugerolle G. 1993. Symbiotic association of Psalteriomonas vulgaris n. spec. with Methanobacterium formicicum. Eur J Protistol 29:98–105. doi: 10.1016/S0932-4739(11)80302-0. [DOI] [PubMed] [Google Scholar]
- 160.Dirren S, Pitsch G, Silva MOD, Posch T. 2017. Grazing of Nuclearia thermophila and Nuclearia delicatula (Nucleariidae, Opisthokonta) on the toxic cyanobacterium Planktothrix rubescens. Eur J Protistol 60:87–101. doi: 10.1016/j.ejop.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 161.Dirren S, Posch T. 2016. Promiscuous and specific bacterial symbiont acquisition in the amoeboid genus Nuclearia (Opisthokonta). FEMS Microbiol Ecol 92:fiw105. doi: 10.1093/femsec/fiw105. [DOI] [PubMed] [Google Scholar]