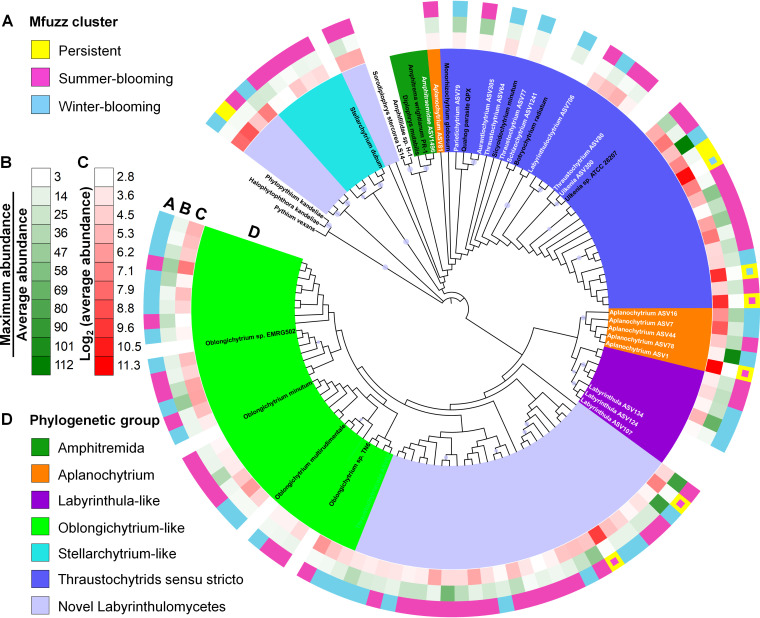
FIG 5.
Maximum likelihood tree constructed using the representative sequences of the 100 most abundant Labyrinthulomycetes 18S rRNA gene amplicon sequence variants (ASVs) and 8 rare but classified ASVs identified from the weekly time series at Piver’s Island Coastal Observatory (PICO) from 2011 to 2013. Reference sequences from NCBI GenBank are included to identify the major clades. The labels of the classified ASVs and reference sequences are indicated by white and black text, respectively. Bootstrap values higher than 80% are indicated by circles. (A to D) The 100 most abundant ASVs are annotated with a number of features, including (A) soft-clustering assignment consistent with Fig. 3, but the persistent ASVs with obvious summer or winter preferences marked with small pink and blue squares, respectively, (B) bloom potential as defined by its maximum relative abundance divided by the average relative abundance across the time series, (C) average relative abundance, and (D) phylogenetic affiliation.