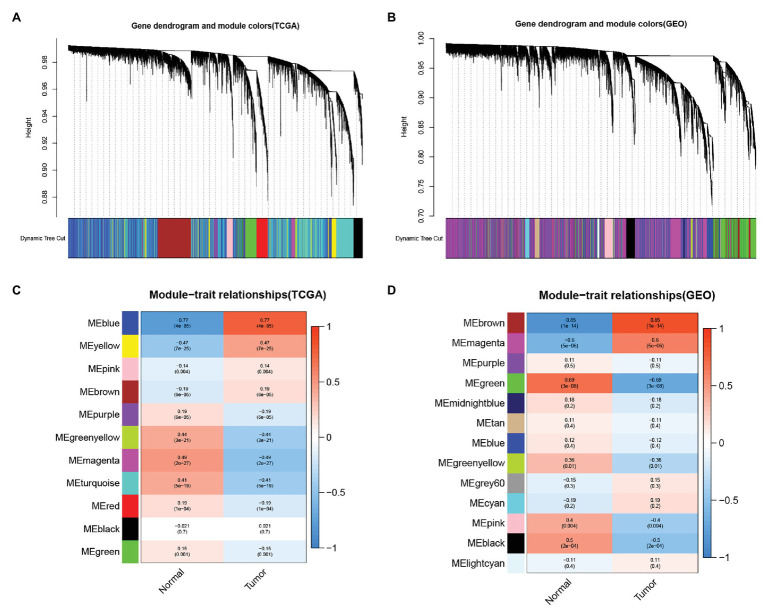
Figure 1.
Identify the module information related to clinical. The clustering tree of the co-expression network module is sorted by hierarchical gene clustering based on the 1-tom matrix. Each module is given a different color. (A) Gene tree and module color in The Cancer Genome Atlas database (TCGA). (B) Gene dendrogram and module color in GSE45114. Module feature relationship. Each row corresponds to a color module, and columns correspond to cancer or normal tissue. Each cell contains the corresponding correlation and value of p. (C) Module trait relationship in TCGA. (D) Module feature relationship inGSE45114.