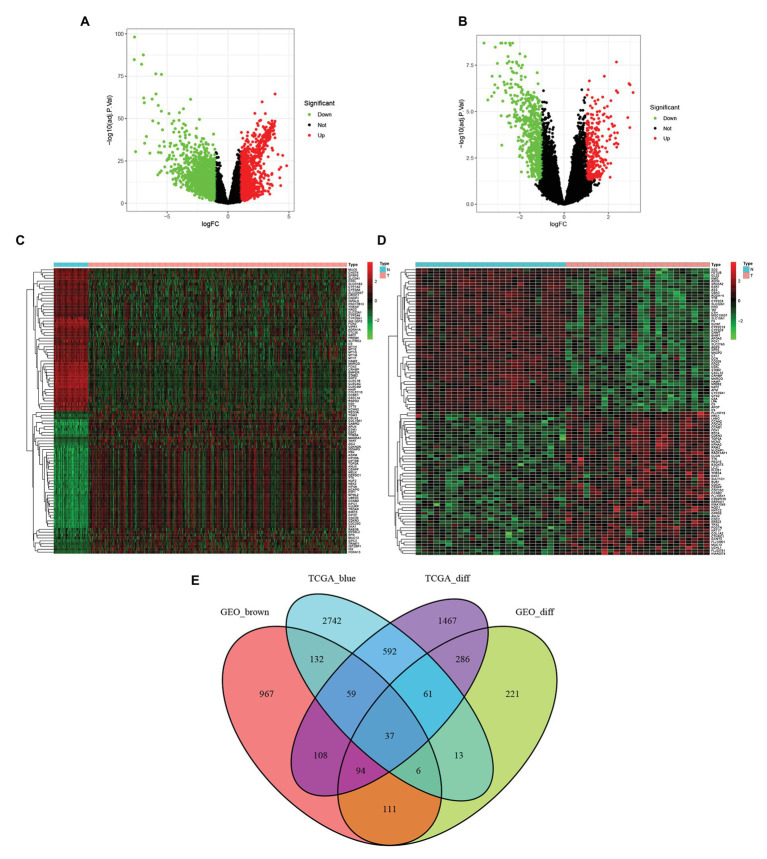
Figure 2.
Using | logfc | > 1.0 and adj p < 0.05 as cut-off criteria, the differentially expressed genes in TCGA and GSE45114 data sets of Hepatocellular carcinoma (HCC) were identified. (A) Volcano maps of differentially expressed genes in the TCGA dataset. (B) Volcano map of differentially expressed genes in the GSE45114 dataset. (C) Heatmap of 100 differentially expressed genes in the TCGA dataset. (D) Heatmap of 100 differential genes in the GSE45114 dataset. (E) Venn diagram of gene crossover between differential expression gene list and co-expression module. A total of 37 overlapping genes were located at the intersection of the differential expression gene list and the co-expression module.