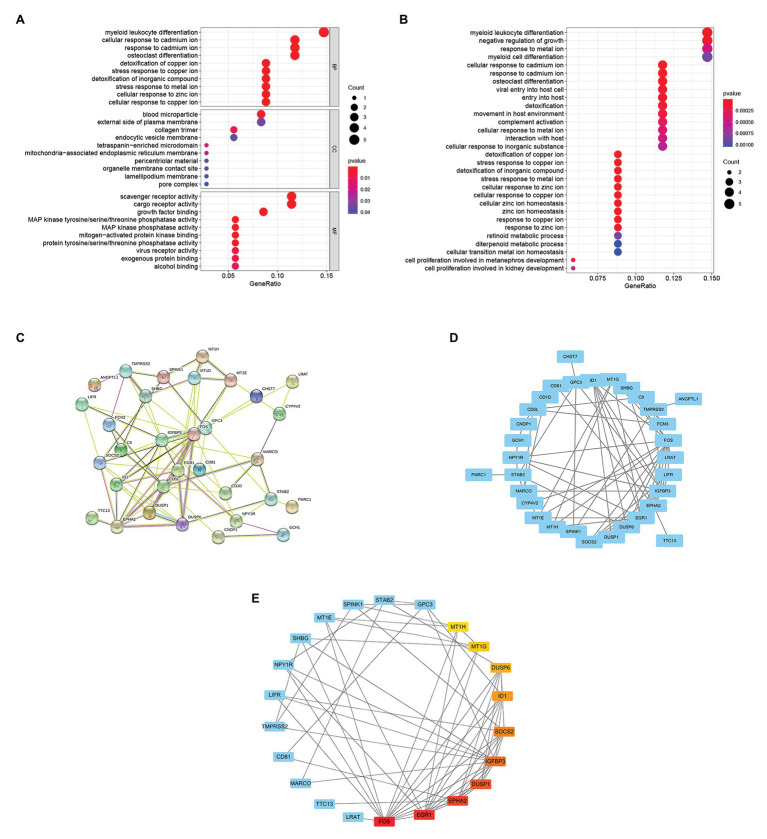
Figure 3.
Enrichment analysis of overlapping genes and selection of hub genes. (A) Gene Ontology (GO) enrichment analysis for the 37 overlapping genes. (B) KEGG pathway enrichment analysis for the 37 overlapping genes. (C) The STRING database constructed the PPI network of 37 overlapping genes. (D) The PPI network of 37 overlapping genes was compiled by Cytoscape software. The blue nodes represent genes, and the edges represent the interaction between nodes. (E) The maximal clique centrality (MCC) algorithm is used to identify hub genes in PPI networks. The edge represents the binding of protein and protein. The red node represents the gene with a high MCC score, while the Yellow node represents the gene with a low MCC score.