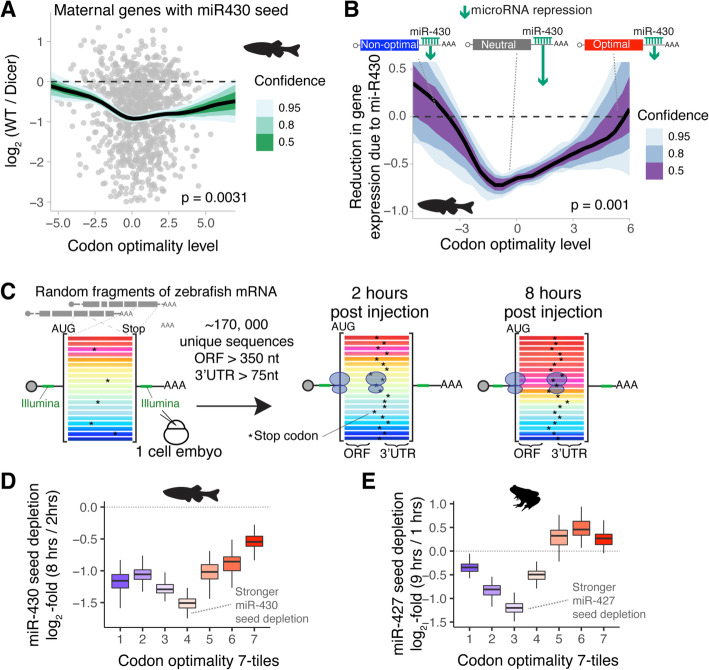
Fig. 5.
Targeting efficacy of miR-430/-427 can be affected by the coding sequence. a Scatter plot comparing codon optimality level and change in expression between wildtype zebrafish embryos vs maternal/zygotic Dicer mutant embryos at 6 h post-fertilization [39]. Only mRNAs with miR-430 seed in the 3′UTR (GCACTT) are shown. The line represents the average change in expression (log2-fold WT/Dicer at 6 hpf) as a function of the codon optimality level (Additional file 1: Table S5). The effect of miR-430 is not constant across different levels of codon optimality (p value was obtained using an F test comparing a model with a non-linear effect on codon optimality vs constant effect). The confidence interval was determined with bootstrap replicates (n = 100) [40]. b Line plot showing the expected decrease in gene expression due to miR-430 during zebrafish MZT (log2-fold change 6 vs 2 hpf) (Additional file 1: Fig. S1a-b). The y-axis represents a measure, estimated from the data, of the miR-430 repressive strength with respect to codon optimality (x-axis). For example, for an mRNA that is very repressed by miR-430, the miR-430 component will be larger (higher negative value in the y-axis). However, for another gene, which has a weak miR-430, the miR-430 component is smaller (closer to 0 in the y-axis). The p value denotes the statistical significance of the non-linear interaction between codon optimality and miR-430 presence (F test) obtained with a generalized additive model [41]. The confidence interval was determined with bootstrap replicates (n = 100) [40]. c Scheme of the reporter library which includes random fragments of the zebrafish transcriptome [22]. Transcripts share the same 5′ and 3′UTR but some sequences contain a stop codon in the coding region. These stop codons create a random and longer 3′UTR sequence. mRNAs were injected at the one‐cell stage in zebrafish [22], and the reporter library is analyzed at 2 and 8 hpi using high-throughput sequencing. To analyze the depletion of miR-430 with respect to the codon content of the transcripts, we filtered those sequences that contain a coding region of at least 350 nucleotides and a random 3′UTR length of at least 75 nucleotides. d, e Analysis of miR-430/427 depletion in the reporter library. The reporters were grouped into 7-tiles, equal size, with increasing levels of codon optimality (Additional file 1: Fig. S5c). For each tile, we computed the depletion of the miR-430/427 seed (GCACTT) in the 3′UTR (log2-fold 8 h/2 h for zebrafish and 9h/1 h for Xenopus). Each boxplot is formed by bootstrap replicates (n = 100) [40]