Abstract
As an important hallmark of metabolic reprogramming in cancer, a disruption in fatty acid metabolism contributes to tumor proliferation, cell migration and invasion, and other tumor cell behaviors. In recent years, more and more studies have been conducted on fatty acid desaturase 2 (FADS2), the first rate-limiting enzyme for the biosynthesis of polyunsaturated fatty acids. These studies have found that FADS2 is abnormally expressed in cancers of the breast, lung, liver, and esophagus; melanoma; leukemia; and other malignant tumors. Furthermore, its expression is significantly correlated with tumor proliferation, cell migration and invasion, clonal formation, angiogenesis, ferroptosis, resistance to radiotherapy, histological grade, metastasis to lymph nodes, clinical stage, and prognosis. The abnormal expression of FADS2 results in an imbalance of cell membrane phospholipids, which disrupts the fluidity of the membrane structure and the transmission of signals and promotes the production of proinflammatory factors and arachidonic acid (AA) metabolites, ultimately harming human health. This article aims to systematically review the structural characteristics of FADS2; its function, expression, and mechanism of action; and the factors affecting its activity. This review also provides new ideas and strategies for the development of treatments aimed at the metabolic reprogramming of tumors.
Keywords: Fatty acid desaturase 2, human cancer, expression, function, metabolism
Introduction
Cancer is the most severe public health problem and the leading cause of death worldwide [1]. There are approximately 4.3 million new cancer cases and 2.9 million new cancer deaths every year in China, and, compared with the United States and the United Kingdom, China has a lower cancer incidence but a 30% and 40% higher cancer mortality [2]. Despite significant advances in the treatment and diagnosis of cancer, the overall survival and prognosis remain poor. Therefore, the identification of new biomarkers and signaling pathways is crucial to the treatment of cancer.
A common phenomenon of tumor cells is metabolic reconstruction, which produces building blocks and energy for tumor cells to grow, divide, and survive. Several studies have reported that disruptions in glycolysis, as well as amino acid (mainly glutamine, serine, and glycine) and lipid metabolism promote the behavior of various malignant tumors by inducing cell proliferation, antiapoptosis, invasion, and metastasis [3], These findings have inspired researchers to explore new strategies for the treatment of various malignant tumors by targeting the metabolism of cancer cells. The breakdown of lipids provides sufficient building blocks and energy for tumor cells to synthesize the cell membrane and to perform other related functions during proliferation [4]. Studies have found that the key enzymes involved in lipid metabolism in tumor cells are acetyl-CoA carboxylase (ACC), fatty acid synthase (FASN), ATP-citrate lyase (ACLY), and fatty acid desaturase (FADS). The dysregulated expression and activity of these key enzymes directly affects the biosynthesis of fatty acids by tumor cells, which in turn interferes with tumor progression [4,5].
As enzymes for fatty acid desaturation, the main function of FADS family is to catalyze the conversion of lipids to regulate the metabolic balance of lipids. In mammals, the FADS family includes FADS1, FADS2, FADS3, FADS4 (SCD5), FADS5 (SCD1), FADS6, FADS7 (DEGS1), and FADS8 (DEGS2). Their amino acid sequences include three common conserved motifs (HX3H, HX2HH and HX2HHXFP), and the differences between them are substrates and sites of action. Among them, SCD has been proven to play an important role in tumor malignant behaviors through the YAP/TAZ pathway, the EGFR/PI3K/AKT pathway, EMT or ferroptosis [6-8], and it has been systematically summarized in several reviews [9,10]. In addition, it has been shown that the common feature of many tumors is increased AA metabolites and increased synthesis of eicosanoids [11]. Omega-6 (n-6) polyunsaturated fatty acids, like arachidonic acid and its metabolites, play an important role in cell activity and physiological functions as components of the microenvironment. Among them, prostaglandin E2, leukotrienes, cyclooxygenase 2, etc. can promote the occurrence and progression of tumors through various mechanisms [12]. FADS2 among the members is a key enzyme that catalyzes the production of such polyunsaturated fatty acids, and changes in the expression and activity of FADS are related to hypertension, metabolic disorders, type 2 diabetes, cardiovascular diseases, inflammation, multiple sclerosis, neurological and mental diseases, and malignancy [13-15]. Although some reports have shown its important functions in cancers, there are few systematic reviews on it. There are fewer studies on other members of the FADS family in cancers. Therefore, the structure and physiological function of FADS2, as well as the role of FADS2 in human cancer, are systematically reviewed in this article.
Gene location and structural characteristics of FADS2
Fatty acid desaturase 2 (FADS2, D6D, des6, llcdl2, fadsd6, tu13, delta-6-desaturase, delta (6) fatty acid desaturase) is encoded by the FADS2 gene on chromosome 11 (11q12-q13.1) [16]. The FADS2 gene has been identified in more than 200 species, including bacteria, fungi, plants, and animals (Table 1) [17-49]. Human FADS2 is a membrane-binding protein of 444 amino acids with a molecular weight of 52.2 kDa; it contains a cytochrome b 5-like domain, two transmembrane domains, and three histidine-rich domains (regions I, II, III). Regions I (HX3H) and II (HX2HH) are located between the two transmembrane domains, and region III (HH) is located at the C-terminus (Figure 1) [18]. FADS2 is expressed in the brain, liver, lungs, heart, and other tissues in humans [18].
Table 1.
FADS2 in different species
| Species name | Amino acid length coded by ORF | Reference |
|---|---|---|
| Cyanobacteria PCC6803 | 359 | [19] |
| Spirulina platensis | 368 | [20] |
| Glossomastix chrysoplasta | 465 | [21] |
| Phaeodactylum tricornutum | 477 | [22] |
| Mortierella alpina | 457/458 | [23-25] |
| Mucor rouxii | 523 | [26] |
| Mucor circinelloides | 467 | [27] |
| Pythium irregulare | 459 | [28] |
| Borago officinalis | 448 | [29] |
| Echium plantagineum | 448 | [30] |
| E. gentianoides and E. pitardii | 438 | [31] |
| Anemone rivularis | 446 | [32] |
| Marchantia polymorpha | 481 | [33] |
| Primula farinosa | 453 | [34] |
| Primula vialii | 453 | [34] |
| Caenorhabditis elegans | 443 | [35] |
| Antheraea pernyi | 316 | [36] |
| Physcomitrella patens | 525 | [37] |
| Ceratodon purpureus | 520 | [38] |
| Oncorhynchus mykiss | 454 | [39] |
| Sparus aurata | 445 | [40] |
| Cyprinus carpio | 445 | [41] |
| Psetta maximus | 445 | [41] |
| Pleuronectiformes | 445 | [41] |
| Cyprinus carpio | 444 | [41,42] |
| Gadus morhua | 447 | [43] |
| Siganus canaliculatus | 445 | [44] |
| Dicentrarchus labrax L. | 445 | [45] |
| Anguilla japonica | 444 | [46] |
| Epinepheluscoioides | 445 | [10] |
| Rachycentron canadum (Linnaeus) | 247 | [9] |
| Eriocheir sinensis | 442 | [47] |
| Mus musculus | 444 | [48] |
| Homo sapiens | 444 | [18] |
| Capra hircus | 444 | [49] |
Figure 1.
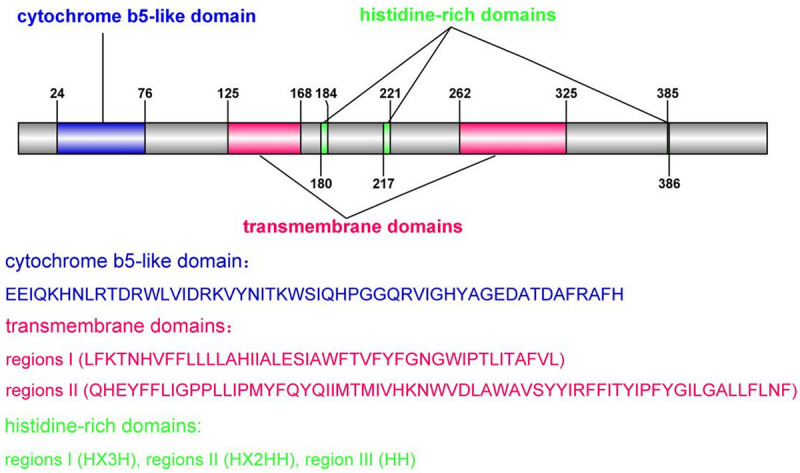
The structure of human FADS2. The blue area is the cytochrome b5-like domain, the red area is the transmembrane domain, and the green area is the histidine-rich domain (region I, II, and III).
Physiological functions of FADS2
FADS2 plays both physiological and pathological roles in different organisms. Wang et al. studied Antheraea pernyi and reported that FADS2 biosynthesis is involved in the mating and communication of these insects [36]. In mice, FADS2 inhibition reduces AA synthesis, thereby reducing inflammation [50], Furthermore, FADS2 gene knockout eliminates the first enzymatic step of the polyunsaturated fatty acid (PUFA) cascade, leading to sexual dysfunction and infertility in male and female mice [51]. Another study has reported that FADS2 regulates the synthesis of PUFAs and the breakdown of triglycerides in dairy goat mammary glands [52]. FADS2 overexpression in zebrafish not only increases the production of PUFAs, but also stimulates antibacterial and anti-inflammatory activity [53].
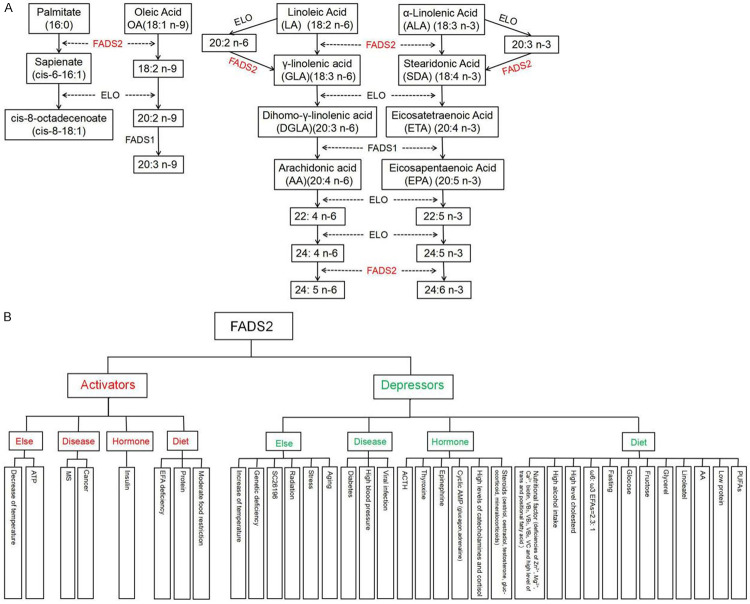
FADS2 mainly plays a role in desaturation by introducing a double bond at the δ6 position of the fatty acid chain, and this is the first rate-limiting enzyme for the conversion of upstream fatty acids into PUFAs (AA and eicosatetraenoic acid). In baboons, a recent study has revealed that in addition to δ6-dehydrogenase activity, FADS2 also possesses δ8-dehydrogenase activity in the desaturation of 20:3n-3 and 20:2n-6 to stearidonic acid (SDA) and γ-linolenic acid (GLA) [54]. FADS2 exhibits desaturase activity toward at least seven substrates (18:2n-6, 18:3n-3, 20:2n-6, 20:3n-3, 24:4n-6, 24:5n-3, 16:0) [55], the major metabolic processes that FADS2 is involved in are shown in Figure 2A. Presently, there is no unified standard for representing the activity of FADS2. Cho et al. used the ratio of ETA/α-linolenic acid (ETA/ALA) and AA/linoleic acid (AA/LA) to represent FADS2 activity [18]. Pender-Cudlip et al. and Gardiner et al. used the ratio of total metabolites of linoleic acid (GLA; DGLA, dihomo-γ-linolenic acid; AA) to LA to express the activity of FADS2 [56,57]. FADS2 activity has also been estimated from the ratio of the percentage of DGLA to the percentage of LA (i.e. % DGLA/% LA) [3] and the ratio of GLA to LA (GLA:LA) [58]. In addition, many factors influence FADS2 activity. These factors are related to different physiological and pathological conditions, such as aging, diet (high alcohol intake; high cholesterol level; deficiencies of zinc, magnesium, and vitamins C, B3, B5, B6; and high trans fatty acid levels), hypertension, diabetes, cardiovascular disease, cancer, viral infections, hormone levels, and allergic dermatitis. FADS2 activity can also be affected by other regulatory genes and single nucleotide polymorphisms, as well as radiation [58-62], (Figure 2B).
Figure 2.
The metabolic pathways catalyzed by FADS2 in organisms (A) and the factors affecting FADS2 activity (B).
Role of FADS2 in cancer
FADS2 and breast cancer
Lane et al. detected FADS2 expression in breast cancer cells and tissues and reported that FADS2 was expressed in the breast cancer cell line MCF-7 but not in the highly invasive triple-negative breast cancer cell MDA-MB-231. In addition, FADS2 expression in breast cancer tissues was significantly lower than that in paracancerous tissues (6.2 ± 2.1 copies/50 ng RNA vs. 15.4 ± 8.2 copies/50 ng RNA, P < 0.05), and it was even lower in breast cancer patients with poor prognoses [63], suggesting that FADS2 is weakly expressed in breast cancer, and low FADS2 expression is significantly correlated with poor prognosis. Vriens et al. found that stearoyl COA destruction (SCD) inhibitor significantly inhibited the proliferation of breast cancer cells MDA-MB-468 and T47D, and FADS2 overexpression restored proliferation. However, FADS2 expression in these two breast cancer cell lines was significantly lower than that in the normal breast epithelial cell line MCF-10A (P < 0.005) [64].
In contrast to the aforementioned results, Zhao et al. analyzed gene expression in human breast cancer and normal tissues by microarray analysis, and the FADS2 mRNA level was more than two times higher in breast cancer tissues than in normal tissues [65]. Pender-Cudlip et al. also showed that FADS2 activity was significantly higher in breast cancer tissues than in paracancerous tissues (1.18 vs. 0.78, P < 0.001), and it was significantly higher in ER− breast cancer tissues than in ER+ breast cancer tissues (1.87 vs. 0.98, P < 0.01), suggesting that the FADS2 expression level may be related to the ER expression level in breast cancer. The discrepancy in the results reported by Lane et al. and others may be attributed to differences in detection methods. Proinflammatory factors, such as PEG2, produced during metabolic processes can alter the inflammatory status of the tumor microenvironment, thereby promoting tumor progression. Pender-Cudlip et al. reported that the amount of PGE2 produced in breast cancer tissues was significantly higher than that in paracancerous tissues (30.81 vs. 6.33 ng/g, P < 0.001), and the amount in ER-breast cancer tissues was significantly higher than that in related paracancerous tissues (P < 0.01). However, in different ER-responsive breast cancer tissues, there was no significant difference in the amount of proinflammatory factors [56], therefore, proinflammatory metabolites are associated with higher FADS2 activity and higher PGE2 expression in more aggressive ER-breast cancer. In conclusion, abnormal FADS2 expression in breast cancer may result in metabolic disorders, which may impact the development and progression of breast cancer.
FADS2 and melanoma
In a mouse model, He et al. reported that FADS2 mRNA and protein levels, as well as its activity, were significantly higher in melanoma B16 tissues than in paracancerous tissues (P < 0.01). FADS2 protein was hardly detected in paracancerous tissues, whereas the FADS2 mRNA level, as well as its activity, were positively correlated with the size of B16 tumors (ETA/LA: P = 0.0015, R2 = 0.58; AA/LA: P = 0.0025, R2 = 0.55; mRNA: R2 = 0.93, P = 0.0017). The content of AA in B16 tumors was four times higher than that in paracancerous tissues, and suppression of FADS2 activity with an inhibitor (SC-26196) or by RNA interference significantly suppressed the growth of B16 tumors (P < 0.01) and significantly reduced the AA content (P < 0.01). Furthermore, protumor metabolite levels derived from AA also decreased by 80%-95%, and the levels of angiogenesis-related genes and inflammatory factors (IL-6 and TNF-α) in B16 tumors were also significantly inhibited. These results indicate that FADS2 is a key factor in the progression of melanoma, and it regulates the release of inflammatory factors and the synthesis of proinflammatory metabolites in the tumor microenvironment [66].
FADS2 and lung cancer
He et al. suggested that FADS2 mRNA and protein levels, as well as activity, were significantly higher in mouse Lewis lung cancer (LCC) tissues than in paracancerous tissues (P < 0.01), and FADS2 protein was undetectable in paracancerous tissues. Furthermore, the FADS2 mRNA level and the enzyme activity were positively correlated with the size of LCC tumors (ETA/LA: P = 0.0001, R2 = 0.70; AA/LA: P = 0.0017, R2 = 0.54; mRNA: P = 0.0139, R2 = 0.81). The content of AA in LCC was two times higher than that in paracancerous tissues, and the suppression of FADS2 activity with an inhibitor (SC-26196) or by RNA interference significantly suppressed the growth of LCC tumors (P < 0.05). Furthermore, the synthesis of AA and AA-derived protumor metabolites in LCC tumors was reduced (P < 0.05), and the levels of angiogenesis-related genes and inflammatory factors were also inhibited [66]. Jiang et al. showed that FADS2 knockdown significantly suppressed lung cancer growth (P < 0.001), thereby resulting in a significant increase in the levels of Fe and lipid reactive oxygen species in lung cancer cells and a significant decrease in the levels of ferroptosis-related genes, which ultimately induces ferroptosis. Survival analysis showed that FADS2 expression is related to the overall survival of lung cancer patients, whereas another mechanistic study found that the expression of LSH in lung cancer positively regulated the expression of the target gene FADS2, and WDR76 increased the expression of FADS2 via epigenetic modifications that require dependence on LSH [67]. Vriens et al. measured the expression of FADS2 in 10 pairs of non-small cell lung cancer/paracancerous tissues and found that the expression of FADS2 in 8 pairs of lung cancer tissues was significantly higher than that in the corresponding paracancerous tissues [64]. In addition to the critical role that FADS2 plays in lung cancer, the circular RNA-circFADS2 produced by abnormal splicing also plays a vital role in lung cancer. Zhao et al. found that the expression of circFADS2 in non-small cell lung cancer tissues and cells was significantly upregulated (P < 0.05), and its high expression was significantly related to poor prognosis, high TNM grade, lymph node metastasis, and poor differentiation of non-small cell lung cancer (P < 0.05). Another study revealed that circFADS2 promotes lung cancer development by regulating miR-498 expression [68]. These findings indicate that the effects of FADS2 on lung cancer cell growth and ferroptosis are regulated by other factors.
FADS2 and brain cancer
Researchers have examined human tissues by microarray and reported that the FADS2 mRNA level in brain tumor tissues was twice as high as that in normal brain tissues [69]. The FADS2 level was also related to the radiotherapy sensitivity of tumors. Wang et al. demonstrated that FADS2 inhibitor SC-26196 decreased the proliferation rate by >45%, decreased the colony formation rate by >40%, and increased the apoptotic rate by 30%-40% for U-87 MG and LN-229 cells under radiotherapy in vitro, and significantly increased the lack of response of tumor growth to radiotherapy in xenograft tumor in mice (66.8 ± 28.3 mm3 vs. 151.6 ± 15.1 mm3). Another study revealed that SC26196 reverses the radioresistance of PGE2-ID1-dependent glioblastoma by blocking the synthesis of AA and PGE2 [70], indicating that FADS2 may be involved in the development of tumor resistance.
FADS2 and liver cancer
Vriens et al. reported that FADS2 mRNA and protein levels are significantly higher in HUH7 liver cancer cells than in normal cells, and the FADS2 level was higher in three out of four pairs of liver cancer tissues than in paracancerous tissues [64]. SCD is a key regulator of various processes such as tumor growth, programmed cell death and carcinogenesis [7]. Vriens et al. also demonstrated that the growth of certain cells was not inhibited when SCD was blocked, and the authors classified the tumor cells as SCD-independent, partially SCD-dependent, and SCD-dependent. The FADS2 level in SCD-independent and partially SCD-dependent tumor cells was significantly higher than that in SCD-dependent tumor cells. In a mechanistic study, liver cancer cells that do not rely on SCD desaturase could synthesize sapienate through FADS2, which is an alternative fatty acid desaturation pathway, to support biofilm synthesis during tumor cell proliferation. This complemented the reduction of fatty acid desaturation caused by the decrease of SCD activity and promoted the proliferation of cancer cells [64], indicating that the FADS2 desaturation pathway plays a key role in promoting the behaviors of certain cancers. However, Horrobin et al. reported no FADS2 expression in human liver cancer [71].
FADS2 and colon cancer
After SC-26196 treatment, tumors in ApcMin/1 mice were reduced by 36%-37%, and the size of primary tumors arising from colon cancer cell HT-29 xenografts in nude mice were reduced by 35% (P < 0.05). Moreover, the LA level in the phospholipids of tissues was significantly increased, and the AA level was decreased. Furthermore, AA supplementation in the diet could eliminate the effects of SC-26196 on the fatty acid composition and tumorigenesis of ApcMin/1 mice, indicating that the effect of this FADS2 inhibitor on the fatty acid composition of these two types of intestinal cancer cell are related to the synthesis of AA [72].
FADS2 and esophageal adenocarcinoma
Wang et al. reported that FADS2 mRNA and protein levels were significantly higher in esophageal adenocarcinoma tissues than in paracancerous tissues (P < 0.05), and the high expression of FADS2 in esophageal adenocarcinoma was significantly correlated with late stage, lymph node metastasis, and poor prognosis (P < 0.001). In vitro, FADS2 overexpression significantly promoted esophageal adenocarcinoma cell proliferation (P < 0.05), and enhanced anchorage- independent colony formation (P < 0.05), and migration and invasion (P < 0.001) [59], indicating that FADS2 may serve as a biomarker of esophageal adenocarcinoma.
FADS2 and other cancers
The results of microarray analysis of human tissues show that the FADS2 mRNA level in cervical cancer tissue is twice as high as that in normal cervical tissue [73]. Vriens et al. reported that FADS2 mRNA and protein levels in the prostate cancer cell line DU145 were significantly higher than those in normal prostate cells [64]. FADS2 activity in renal cell carcinoma was significantly higher than that in healthy renal tissue [74]. Apart from the aforementioned solid tumors, Agatha et al. found a significant increase in FADS2 indexes of membrane phospholipids in children with acute lymphoblastic leukemia (1.21 ± 0.39 vs. 0.27 ± 0.04; P < 0.001). However, there was no significant difference in FADS2 indexes of membrane phospholipids in children with acute myelogenous leukemia (0.26 ± 0.08 vs. 0.27 ± 0.04). FADS2 activity related to n-6 and n-3 pathways in blood cells of patients with acute lymphoblastic leukemia increased by 3.8-fold and 2.5-fold compared to the healthy control group, respectively. However, there was no significant change in FADS2 activity in blood cells of patients with acute myelogenous leukemia [75]. In addition to the abnormal expression in leukemia cells, inhibition of FADS2 could suppress the growth of various leukemia cells [76]. These data indicate that changes in FADS2 expression and activity in cervical cancer, prostate cancer, renal carcinoma, and acute lymphoblastic leukemia may be related to the development and progression of tumors.
Discussion and conclusion
The studies reviewed in this article indicate that FADS2 overexpression promotes tumor proliferation, clonal formation, migration and invasion, and lymph node metastasis, and it is related to the tumor microenvironment, poor prognosis, radiotherapy resistance, and ferroptosis. However, some researchers have reported that inhibition of the FADS2 level in tumor cells in vitro cannot suppress the proliferation of tumor cells. For example, in the study conducted by He et al., tumor cell growth was not suppressed after inhibition of FADS2 by SC-26196 and RNA interference in B16 melanoma and LCC lung cancer, which may be attributed to the fact that inhibiting FADS2 is not directly toxic to tumor cells. Instead, it may affect the behavior of tumor cells by regulating the production of metabolites in the tumor microenvironment or altering the phospholipid composition of the tumor cell membrane [66]. Vriens et al. indicated that silencing FADS2 in HUH7 cells promotes proliferation, which may be the result of an alternative pathway of FADS2 fatty acid metabolism in tumor cells [64]. This article also summarizes studies in which FADS2 activity was up- or downregulated, and studies in which the expression in certain cancers, such as breast cancer and liver cancer, resulted in different conclusions (Table 2). Discrepancies may be related to the timespan between specific studies and available detection methods at the times they were performed.
Table 2.
The association between FADS2 expression and the malignant tumors and its functions
| Cancer type | FADS2 expression level | Function | Reference |
|---|---|---|---|
| Breast cancer | Downregulated (mRNA and activity) | Low FADS2 expression was related to poor prognosis of breast cancer patients and high TNM grade of tumor. | [63,64,77] |
| Upregulated (mRNA and activity) | High expression promoted the synthesis of the pro-inflammatory metabolite PGE2 in breast cancer, which may promote the occurrence of inflammation. | [56,65] | |
| Melanoma | Upregulated (mRNA, protein and activity) | High expression promoted the growth of B16 melanoma and could lead to enhanced n-6 AA and AA-derived tumor, promoting metabolites production, as well as the gene expression of angiogenesis and inflammatory factors. | [66] |
| Lung cancer | Upregulated (mRNA, protein and activity) | High expression promoted the growth of lung cancer and could lead to enhanced n-6 AA and AA-derived tumor, promoting metabolites production, as well as the gene expression of angiogenesis and inflammatory factors. Inhibiting FADS2 could induce ferroptosis by increasing the level of Fe and lipid ROS in lung cancer cells. Patients with high FADS2 expression had poorer prognosis. | [64,66,67] |
| Brain cancer | Upregulated (mRNA) | Inhibition of FADS2 could improve the sensitivity of tumor cells to radiotherapy and block the synthesis of AA and PGE2 in vitro and in vivo. | [69,70] |
| Liver cancer | Loss | -- | [71] |
| Upregulated (mRNA and protein) | FADS2 promoted the proliferation of cancer cells by synthesizing sapienate and used it for membrane biosynthesis of cancer cell. | [64] | |
| Colon cancer | -- | Inhibition of FADS2 could inhibit the growth of colon cancer and the synthesis of AA. | [72] |
| Esophageal carcinoma | Upregulated (mRNA and protein) | High expression was related to clinicopathological parameters such as late stage, lymph node metastasis, and poor prognosis. High expression promoted tumor cell proliferation, non-anchored clonal formation, migration and invasion. | [59] |
| Cervical cancer | Upregulated (mRNA) | -- | [73] |
| Prostate cancer | Upregulated (mRNA and protein) | -- | [64] |
| Renal cell carcinoma | Increased (activity) | -- | [74] |
| Acute lymphoblastic leukemia | Increased (activity) | High activity promoted the growth of different types of leukemia cells. | [75,76] |
Thus far, there are two main mechanisms of action of FADS2 in cancer: (1) Metabolite-related mechanisms in which FADS2 modulates the tumor microenvironment or the fluidity of cell membranes by regulating the synthesis of PUFAs during fatty acid metabolism. For example, Scanferlato et al. found that the production of sapienic acid is related to apoptosis and necrosis in colon cancer cells CaO2 [78]. Furthermore, FADS2 supports the synthesis of cell membranes during tumor cell proliferation by catalyzing the conversion of palmitic acid to sapienate and its extended product, cis-8-octadecenoic acid [64]. FADS2 inhibitors can reverse PGE2-ID1-dependent radioresistance in glioblastoma cells by blocking the synthesis of AA and PGE2 [70]. Pender-Cudlip et al. suggest that the AA synthesis pathway (AA and PGE2 production), which involves FADS2, may be related to breast cancer [56]. He et al. demonstrate that FADS2 may impair the tumor microenvironment by regulating the synthesis of AA and AA-derived eicosanoids (PGs, LTs, EETs), and thus affects tumor growth [66]. PUFAs can also regulate the expression of various transcription factors, including PPAR α/β/γ1/γ2, SREBP-1c, HNF-4α/γ, RXRα, LXRα, and NF-κB by directly binding transcription factors or regulating signal transduction pathways that control expression, phosphorylation, ubiquitination, or proteolysis in the liver [79]. Furthermore, AA-derived products such as prostaglandins, thromboxane, leukenoic acid, and 5-hydroxyeicosapentaenoic acid play important roles in cancer and other diseases [12]. (2) Molecular regulation-related mechanisms in which FADS2 expression or activity is regulated by other factors. Wy14643, an activator of transcription factor PPARα, synergistically induces FADS2 transcription [80]. WDR76 and WD40 proteins can increase the expression of FADS2 through the epigenetic modification of the TSSs of the FADS2 promoter [67]. SREBP-1 and PPAR-α are involved in fatty acid biosynthesis by regulating FADS2 transcription [81,82]. As a key transcription factor in fatty acid metabolism, SREBP can be activated by mTOR signaling [83], and HIF-1α can stimulate SREBP-1c expression under hypoxia [84]. PUFAs, metabolites of FADS2, can also regulate the nuclear abundance of SREBP-1 and PPARα in liver and regulate the expression of FADS2 through a feedback mechanism [85,86]. He et al. hypothesized the possible molecular regulatory mechanism of FADS2 expression in cancer, namely, hypoxia/reactive oxygen species-HIF-1α-SREBP-1c-FADS2 [66]. Recently, a research showed that SREBP-1/2 and mTOR signaling can regulate the expression of FADS2 and the production of its metabolite sapienate in cancer cells, just as we guessed [87]. The molecular mechanism of FADS2 in tumors needs further experimental exploration.
This review illustrates the important roles of FADS2 in the development, progression, metabolism, and death of various cancer cells. However, in studies on the relationship between FADS2 and cancer, the mechanisms by which FADS2 regulates tumor cells are rarely addressed. Thus, the expression, function, and specific mechanism of FADS2 in specific cancers need to be further explored. The regulation of FADS2 may have potential anti-inflammatory and antitumor effects in different pathological conditions. Therefore, it is important to explore the association between FADS2 and other clinicopathological factors in cancer patients such as age, lifestyle, menopause status, histological grade, and genotypes. Drug research and nutritional interventions for the breakdown of fatty acids, which is catalyzed by FADS2, may provide new strategies for cancer prevention and treatment.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (No. 81102030), Biotechnology Innovation Program of Southwest Hospital (No. SWH2016JCYB-39), Talents Training Program of Third Military Medical University (No. 2017MPRC-18), Chongqing Basic Research and Frontier Exploration Project (No. cstc2018jcyjA0137) and Military Medical Staff Innovation Plan of Southwest Hospital (No. SWH2018BJLC-04) as well as Army Medical University (No. XZ-2019-505-042).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 2.Feng RM, Zong YN, Cao SM, Xu RH. Current cancer situation in China: good or bad news from the 2018 Global Cancer Statistics? Cancer Commun (Lond) 2019;39:22. doi: 10.1186/s40880-019-0368-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Z, Zhang H. Reprogramming of glucose, fatty acid and amino acid metabolism for cancer progression. Cell Mol Life Sci. 2016;73:377–392. doi: 10.1007/s00018-015-2070-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ray U, Roy SS. Aberrant lipid metabolism in cancer cells - the role of oncolipid-activated signaling. FEBS J. 2018;285:432–443. doi: 10.1111/febs.14281. [DOI] [PubMed] [Google Scholar]
- 5.Koundouros N, Poulogiannis G. Reprogramming of fatty acid metabolism in cancer. Br J Cancer. 2020;122:4–22. doi: 10.1038/s41416-019-0650-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noto A, De Vitis C, Pisanu ME, Roscilli G, Ricci G, Catizone A, Sorrentino G, Chianese G, Taglialatela-Scafati O, Trisciuoglio D, Del Bufalo D, Di Martile M, Di Napoli A, Ruco L, Costantini S, Jakopin Z, Budillon A, Melino G, Del Sal G, Ciliberto G, Mancini R. Stearoyl-CoA-desaturase 1 regulates lung cancer stemness via stabilization and nuclear localization of YAP/TAZ. Oncogene. 2017;36:4573–4584. doi: 10.1038/onc.2017.75. [DOI] [PubMed] [Google Scholar]
- 7.She K, Fang S, Du W, Fan X, He J, Pan H, Huang L, He P, Huang J. SCD1 is required for EGFR-targeting cancer therapy of lung cancer via re-activation of EGFR/PI3K/AKT signals. Cancer Cell Int. 2019;19:103. doi: 10.1186/s12935-019-0809-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tesfay L, Paul BT, Konstorum A, Deng Z, Cox AO, Lee J, Furdui CM, Hegde P, Torti FM, Torti SV. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 2019;79:5355–5366. doi: 10.1158/0008-5472.CAN-19-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Igal RA. Roles of StearoylCoA Desaturase-1 in the regulation of cancer cell growth, survival and tumorigenesis. Cancers (Basel) 2011;3:2462–2477. doi: 10.3390/cancers3022462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tracz-Gaszewska Z, Dobrzyn P. Stearoyl-CoA Desaturase 1 as a therapeutic target for the treatment of cancer. Cancers. 2019;11:948. doi: 10.3390/cancers11070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mazhar D, Ang R, Waxman J. COX inhibitors and breast cancer. Br J Cancer. 2006;94:346–350. doi: 10.1038/sj.bjc.6602942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomes RN, Felipe da Costa S, Colquhoun A. Eicosanoids and cancer. Clinics (Sao Paulo) 2018;73:e530s. doi: 10.6061/clinics/2018/e530s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosi F, Sartori F, Guarini P, Olivieri O, Martinelli N. Delta-5 and delta-6 desaturases: crucial enzymes in polyunsaturated fatty acid-related pathways with pleiotropic influences in health and disease. Adv Exp Med Biol. 2014;824:61–81. doi: 10.1007/978-3-319-07320-0_7. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, Jandacek R, Rider T, Tso P, McNamara RK. Elevated delta-6 desaturase (FADS2) expression in the postmortem prefrontal cortex of schizophrenic patients: relationship with fatty acid composition. Schizophr Res. 2009;109:113–120. doi: 10.1016/j.schres.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, McNamara RK. Elevated delta-6 desaturase (FADS2) gene expression in the prefrontal cortex of patients with bipolar disorder. J Psychiatr Res. 2011;45:269–272. doi: 10.1016/j.jpsychires.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura MT, Nara TY. Structure, function, and dietary regulation of Δ6, Δ5, and Δ9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- 17.Yang ZG, Guo ZH, Yao QQ, Cheng YX. Research advance in fatty acid desaturase genes. Biotechnol Bull. 2013;12:21–26. [Google Scholar]
- 18.Cho HP, Nakamura MT, Clarke SD. Cloning, expression, and nutritional regulation of the mammalian Delta-6 desaturase. J Biol Chem. 1999;274:471–477. doi: 10.1074/jbc.274.1.471. [DOI] [PubMed] [Google Scholar]
- 19.Reddy AS, Nuccio ML, Gross LM, Thomas TL. Isolation of a delta 6-desaturase gene from the cyanobacterium Synechocystis sp. strain PCC 6803 by gain-of-function expression in Anabaena sp. strain PCC 7120. Plant Mol Biol. 1993;22:293–300. doi: 10.1007/BF00014936. [DOI] [PubMed] [Google Scholar]
- 20.Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, Widhalm K, Moschonis G, Stehle P, Amouyel P, De Henauw S, Molnàr D, Moreno LA, Meirhaeghe A, Dallongeville J. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51:2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subudhi S, Kurdrid P, Hongsthong A, Sirijuntarut M, Cheevadhanarak S, Tanticharoen M. Isolation and functional characterization of Spirulina D6D gene promoter: role of a putative GntR transcription factor in transcriptional regulation of D6D gene expression. Biochem Biophys Res Commun. 2008;365:643–649. doi: 10.1016/j.bbrc.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Domergue F, Lerchl J, Zähringer U, Heinz E. Cloning and functional characterization of Phaeodactylum tricornutum front-end desaturases involved in eicosapentaenoic acid biosynthesis. Eur J Biochem. 2002;269:4105–4113. doi: 10.1046/j.1432-1033.2002.03104.x. [DOI] [PubMed] [Google Scholar]
- 23.Huang YS, Chaudhary S, Thurmond JM, Bobik EG Jr, Yuan L, Chan GM, Kirchner SJ, Mukerji P, Knutzon DS. Cloning of delta12- and delta6-desaturases from Mortierella alpina and recombinant production of gamma-linolenic acid in Saccharomyces cerevisiae. Lipids. 1999;34:649–659. doi: 10.1007/s11745-999-0410-8. [DOI] [PubMed] [Google Scholar]
- 24.Sakuradani E, Shimizu S. Gene cloning and functional analysis of a second delta 6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Biosci Biotechnol Biochem. 2003;67:704–711. doi: 10.1271/bbb.67.704. [DOI] [PubMed] [Google Scholar]
- 25.Shi H, Chen H, Gu Z, Zhang H, Chen W, Chen YQ. Application of a delta-6 desaturase with alpha-linolenic acid preference on eicosapentaenoic acid production in Mortierella alpina. Microb Cell Fact. 2016;15:117. doi: 10.1186/s12934-016-0516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laoteng K, Mannontarat R, Tanticharoen M, Cheevadhanarak S. Delta(6)-desaturase of Mucor rouxii with high similarity to plant delta(6)-desaturase and its heterologous expression in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2000;279:17–22. doi: 10.1006/bbrc.2000.3856. [DOI] [PubMed] [Google Scholar]
- 27.Hao YL, Mei XH, Zhao F, Yin S, Li RH, Luo YB. Expression of Mucor circinelloides gene for Δ6 desaturase results in the accumulation of high levels of γ-linolenic acid in transgenic tobacco. Russ J Plant Physiol. 2011;55:77–81. [Google Scholar]
- 28.Hong H, Datla N, Reed DW, Covello PS, MacKenzie SL, Qiu X. High-level production of gamma-linolenic acid in Brassica juncea using a delta6 desaturase from Pythium irregulare. Plant Physiol. 2002;129:354–362. doi: 10.1104/pp.001495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sayanova O, Smith MA, Lapinskas P, Stobart AK, Dobson G, Christie WW, Shewry PR, Napier JA. Expression of a borage desaturase cDNA containing an N-terminal cytochrome b5 domain results in the accumulation of high levels of delta6-desaturated fatty acids in transgenic tobacco. Proc Natl Acad Sci U S A. 1997;94:4211–4216. doi: 10.1073/pnas.94.8.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou XR, Robert S, Singh S, Green A. Heterologous production of GLA and SDA by expression of an Echium plantagineum Δ6-desaturase gene. Plant Sci. 2006;170:665–673. [Google Scholar]
- 31.Garcia-Maroto F, Garrido-Cardenas JA, Rodriguez-Ruiz J, Vilches-Ferron M, Adam AC, Polaina J, Alonso DL. Cloning and molecular characterization of the delta6-desaturase from two echium plant species: production of GLA by heterologous expression in yeast and tobacco. Lipids. 2002;37:417–426. doi: 10.1007/s1145-002-0910-6. [DOI] [PubMed] [Google Scholar]
- 32.Whitney HM, Michaelson LV, Sayanova O, Pickett JA, Napier JA. Functional characterisation of two cytochrome b5-fusion desaturases from Anemone leveillei: the unexpected identification of a fatty acid Delta6-desaturase. Planta. 2003;217:983–992. doi: 10.1007/s00425-003-1069-5. [DOI] [PubMed] [Google Scholar]
- 33.Kajikawa M, Yamato KT, Kohzu Y, Nojiri M, Sakuradani E, Shimizu S, Sakai Y, Fukuzawa H, Ohyama K. Isolation and characterization of delta(6)-desaturase, an ELO-like enzyme and delta(5)-desaturase from the liverwort Marchantia polymorpha and production of arachidonic and eicosapentaenoic acids in the methylotrophic yeast Pichia pastoris. Plant Mol Biol. 2004;54:335–352. doi: 10.1023/B:PLAN.0000036366.57794.ee. [DOI] [PubMed] [Google Scholar]
- 34.Sayanova OV, Beaudoin F, Michaelson LV, Shewry PR, Napier JA. Identification of primula fatty acid delta 6-desaturases with n-3 substrate preferences. FEBS Lett. 2003;542:100–104. doi: 10.1016/s0014-5793(03)00358-2. [DOI] [PubMed] [Google Scholar]
- 35.Napier JA, Hey SJ, Lacey DJ, Shewry PR. Identification of a Caenorhabditis elegans Delta6-fatty-acid-desaturase by heterologous expression in Saccharomyces cerevisiae. Biochem J. 1998;330:611–614. doi: 10.1042/bj3300611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang HL, Liénard MA, Zhao CH, Wang CZ, Löfstedt C. Neofunctionalization in an ancestral insect desaturase lineage led to rare Δ6 pheromone signals in the Chinese tussah silkworm. Insect Biochem Mol Biol. 2010;40:742–751. doi: 10.1016/j.ibmb.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Girke T, Schmidt H, Zahringer U, Reski R, Heinz E. Identification of a novel delta 6-acyl-group desaturase by targeted gene disruption in Physcomitrella patens. Plant J. 1998;15:39–48. doi: 10.1046/j.1365-313x.1998.00178.x. [DOI] [PubMed] [Google Scholar]
- 38.Sperling P, Lee M, Girke T, Zahringer U, Stymne S, Heinz E. A bifunctional delta-fatty acyl acetylenase/desaturase from the moss Ceratodon purpureus. A new member of the cytochrome b5 superfamily. Eur J Biochem. 2000;267:3801–3811. doi: 10.1046/j.1432-1327.2000.01418.x. [DOI] [PubMed] [Google Scholar]
- 39.Seiliez I, Panserat S, Kaushik S, Bergot P. Cloning, tissue distribution and nutritional regulation of a Delta6-desaturase-like enzyme in rainbow trout. Comp Biochem Physiol B Biochem Mol Biol. 2001;130:83–93. doi: 10.1016/s1096-4959(01)00410-9. [DOI] [PubMed] [Google Scholar]
- 40.Seiliez I, Panserat S, Corraze G, Kaushik S, Bergot P. Cloning and nutritional regulation of a Delta6-desaturase-like enzyme in the marine teleost gilthead seabream (Sparus aurata) Comp Biochem Physiol B Biochem Mol Biol. 2003;135:449–460. doi: 10.1016/s1096-4959(03)00111-8. [DOI] [PubMed] [Google Scholar]
- 41.Zheng X, Seiliez I, Hastings N, Tocher DR, Panserat S, Dickson CA, Bergot P, Teale AJ. Characterization and comparison of fatty acyl Δ6 desaturase cDNAs from freshwater and marine teleost fish species. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:269–279. doi: 10.1016/j.cbpc.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Ren HT, Yu JH, Xu P, Tang YK. Influence of dietary fatty acids on muscle fatty acid composition and expression levels of Delta6 desaturase-like and Elovl5-like elongase in common carp (Cyprinus carpio var. Jian) Comp Biochem Physiol B Biochem Mol Biol. 2012;163:184–192. doi: 10.1016/j.cbpb.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 43.Tocher DR, Zheng X, Schlechtriem C, Hastings N, Dick JR, Teale AJ. Highly unsaturated fatty acid synthesis in marine fish: cloning, functional characterization, and nutritional regulation of fatty acyl delta 6 desaturase of Atlantic cod (Gadus morhua L.) Lipids. 2006;41:1003–1016. doi: 10.1007/s11745-006-5051-4. [DOI] [PubMed] [Google Scholar]
- 44.Hu CB. Cloning of delta 6 desaturase cDNA and effects of ambient salinity and dietary lipids on its mRNA expression in rabbitfish, Siganus Oramin. Shantou University. 2007 [Google Scholar]
- 45.González-Rovira A, Mourente G, Zheng X, Tocher DR, Pendón C. Molecular and functional characterization and expression analysis of a Δ6 fatty acyl desaturase cDNA of European Sea Bass (Dicentrarchus labrax L.) Aquaculture. 2009;298:90–100. [Google Scholar]
- 46.Kikuchi K, Tsukamoto H. Stearoyl-CoA desaturase and tumorigenesis. Chem Biol Interact. 2020;316:108917. doi: 10.1016/j.cbi.2019.108917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang Z, Guo Z, Ji L, Zeng Q, Wang Y, Yang X, Cheng Y. Cloning and tissue distribution of a fatty acyl Delta6-desaturase-like gene and effects of dietary lipid levels on its expression in the hepatopancreas of Chinese mitten crab (Eriocheir sinensis) Comp Biochem Physiol B Biochem Mol Biol. 2013;165:99–105. doi: 10.1016/j.cbpb.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Aki T, Shimada Y, Inagaki K, Higashimoto H, Kawamoto S, Shigeta S, Ono K, Suzuki O. Molecular cloning and functional characterization of rat delta-6 fatty acid desaturase. Biochem Biophys Res Commun. 1999;255:575–579. doi: 10.1006/bbrc.1999.0235. [DOI] [PubMed] [Google Scholar]
- 49.Wu JHQ, Li Z, Li C, Wang H, Shi HP, Luo J. Effects of FADS2 interference on fatty acid composition of mammary epithelial cells in dairy goats (Capra hircus) J Agric Biotechnol. 2019;27:1973–1984. [Google Scholar]
- 50.Franco P, De Rose F, De Santis MC, Pasinetti N, Lancellotta V, Meduri B, Meattini I Clinical Oncology Breast Cancer Group (COBCG) Investigators. Omission of postoperative radiation after breast conserving surgery: a progressive paradigm shift towards precision medicine. Clin Transl Radiat Oncol. 2020;21:112–119. doi: 10.1016/j.ctro.2020.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoffel W, Holz B, Jenke B, Binczek E, Gunter RH, Kiss C, Karakesisoglou I, Thevis M, Weber AA, Arnhold S, Addicks K. Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 2008;27:2281–2292. doi: 10.1038/emboj.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, He QY, Li Z, Li C, Wang H, Shi HP, Luo J. Effects of FADS2 interference on fatty acid composition of mammary epithelial cells in dairy goats (Capra hircus) J Agric Biotechnol. 2019;27:1973–1984. [Google Scholar]
- 53.Wang YD, Peng KC, Wu JL, Chen JY. Transgenic expression of salmon delta-5 and delta-6 desaturase in zebrafish muscle inhibits the growth of Vibrio alginolyticus and affects fish immunomodulatory activity. Fish Shellfish Immunol. 2014;39:223–230. doi: 10.1016/j.fsi.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 54.Park WJ, Kothapalli KS, Lawrence P, Tyburczy C, Brenna JT. An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J Lipid Res. 2009;50:1195–1202. doi: 10.1194/jlr.M800630-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Park WJ, Kothapalli KS, Lawrence P, Brenna JT. FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS One. 2011;6:e28186. doi: 10.1371/journal.pone.0028186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pender-Cudlip MC, Krag KJ, Martini D, Yu J, Guidi A, Skinner SS, Zhang Y, Qu X, He C, Xu Y, Qian SY, Kang JX. Delta-6-desaturase activity and arachidonic acid synthesis are increased in human breast cancer tissue. Cancer Sci. 2013;104:760–764. doi: 10.1111/cas.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rezapour-Firouzi S, Arefhosseini SR, Farhoudi M, Ebrahimi-Mamaghani M, Rashidi MR, Torbati MA, Baradaran B. Association of expanded disability status scale and cytokines after intervention with co-supplemented hemp seed, evening primrose oils and hot-natured diet in multiple sclerosis patients(diamond) Bioimpacts. 2013;3:43–47. doi: 10.5681/bi.2013.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Arshad Z, Rezapour-Firouzi S, Ebrahimifar M, Mosavi Jarrahi A, Mohammadian M. Association of delta-6-desaturase expression with aggressiveness of cancer, diabetes mellitus, and multiple sclerosis: a narrative review. Asian Pac J Cancer Prev. 2019;20:1005–1018. doi: 10.31557/APJCP.2019.20.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lattka E, Eggers S, Moeller G, Heim K, Weber M, Mehta D, Prokisch H, Illig T, Adamski J. A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res. 2010;51:182–191. doi: 10.1194/jlr.M900289-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brenner RR. Nutritional and hormonal factors influencing desaturation of essential fatty acids. Prog Lipid Res. 1981;20:41–47. doi: 10.1016/0163-7827(81)90012-6. [DOI] [PubMed] [Google Scholar]
- 61.Brenner RR. Endocrine control of fatty acid desaturation. Biochem Soc Trans. 1990;18:773–775. doi: 10.1042/bst0180773. [DOI] [PubMed] [Google Scholar]
- 62.Horrobin DF. Nutritional and medical importance of gamma-linolenic acid. Prog Lipid Res. 1992;31:163–194. doi: 10.1016/0163-7827(92)90008-7. [DOI] [PubMed] [Google Scholar]
- 63.Lane J, Mansel RE, Jiang WG. Expression of human delta-6-desaturase is associated with aggressiveness of human breast cancer. Int J Mol Med. 2003;12:253–257. [PubMed] [Google Scholar]
- 64.Vriens K, Christen S, Parik S, Broekaert D, Yoshinaga K, Talebi A, Dehairs J, Escalona-Noguero C, Schmieder R, Cornfield T, Charlton C, Romero-Perez L, Rossi M, Rinaldi G, Orth MF, Boon R, Kerstens A, Kwan SY, Faubert B, Mendez-Lucas A, Kopitz CC, Chen T, Fernandez-Garcia J, Duarte JAG, Schmitz AA, Steigemann P, Najimi M, Hagebarth A, Van Ginderachter JA, Sokal E, Gotoh N, Wong KK, Verfaillie C, Derua R, Munck S, Yuneva M, Beretta L, DeBerardinis RJ, Swinnen JV, Hodson L, Cassiman D, Verslype C, Christian S, Grunewald S, Grunewald TGP, Fendt SM. Evidence for an alternative fatty acid desaturation pathway increasing cancer plasticity. Nature. 2019;566:403–406. doi: 10.1038/s41586-019-0904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, Jeffrey SS. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–2536. doi: 10.1091/mbc.E03-11-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.He C, Qu X, Wan J, Rong R, Huang L, Cai C, Zhou K, Gu Y, Qian SY, Kang JX. Inhibiting delta-6 desaturase activity suppresses tumor growth in mice. PLoS One. 2012;7:e47567. doi: 10.1371/journal.pone.0047567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jiang Y, Mao C, Yang R, Yan B, Shi Y, Liu X, Lai W, Liu Y, Wang X, Xiao D, Zhou H, Cheng Y, Yu F, Cao Y, Liu S, Yan Q, Tao Y. EGLN1/c-Myc induced lymphoid-specific helicase inhibits ferroptosis through lipid metabolic gene expression changes. Theranostics. 2017;7:3293–3305. doi: 10.7150/thno.19988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao F, Han Y, Liu Z, Zhao Z, Li Z, Jia K. circFADS2 regulates lung cancer cells proliferation and invasion via acting as a sponge of miR-498. Biosci Rep. 2018;38:BSR20180570. doi: 10.1042/BSR20180570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, Rosenblum M, Mikkelsen T, Fine HA. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, Liang H, Sun M, Zhang L, Xu H, Liu W, Li Y, Zhou Y, Li Y, Li M. Delta-6-desaturase inhibitor enhances radiation therapy in glioblastoma in vitro and in vivo. Cancer Manag Res. 2018;10:6779–6790. doi: 10.2147/CMAR.S185601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Horrobin DF. The reversibility of cancer: the relevance of cyclic AMP, calcium, essential fatty acids and prostaglandin E1. Med Hypotheses. 1980;6:469–486. doi: 10.1016/0306-9877(80)90099-7. [DOI] [PubMed] [Google Scholar]
- 72.Hansen-Petrik MB, McEntee MF, Johnson BT, Obukowicz MG, Masferrer J, Zweifel B, Chiu CH, Whelan J. Selective inhibition of Delta-6 desaturase impedes intestinal tumorigenesis. Cancer Lett. 2002;175:157–163. doi: 10.1016/s0304-3835(01)00715-7. [DOI] [PubMed] [Google Scholar]
- 73.Scotto L, Narayan G, Nandula SV, Arias-Pulido H, Subramaniyam S, Schneider A, Kaufmann AM, Wright JD, Pothuri B, Mansukhani M, Murty VV. Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression. Genes Chromosomes Cancer. 2008;47:755–765. doi: 10.1002/gcc.20577. [DOI] [PubMed] [Google Scholar]
- 74.Hoffmann K, Blaudszun J, Brunken C, Hopker WW, Tauber R, Steinhart H. Distribution of polyunsaturated fatty acids including conjugated linoleic acids in total and subcellular fractions from healthy and cancerous parts of human kidneys. Lipids. 2005;40:309–315. doi: 10.1007/s11745-005-1387-z. [DOI] [PubMed] [Google Scholar]
- 75.Agatha G, Hafer R, Zintl F. Fatty acid composition of lymphocyte membrane phospholipids in children with acute leukemia. Cancer Lett. 2001;173:139–144. doi: 10.1016/s0304-3835(01)00674-7. [DOI] [PubMed] [Google Scholar]
- 76.Agatha G, Voigt A, Kauf E, Zintl F. Conjugated linoleic acid modulation of cell membrane in leukemia cells. Cancer Letters. 2004;209:87–103. doi: 10.1016/j.canlet.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 77.Grammatikos S, Harvey M, Subbaiah P, Victor T, Miller W. Loss of fatty-acid delta(6) desaturating ability in human mammary epithelial-cells that express an activated C-ha-ras oncogene. Int J Oncol. 1995;6:1039–1046. doi: 10.3892/ijo.6.5.1039. [DOI] [PubMed] [Google Scholar]
- 78.Scanferlato R, Bortolotti M, Sansone A, Chatgilialoglu C, Polito L, De Spirito M, Maulucci G, Bolognesi A, Ferreri C. Hexadecenoic fatty acid positional isomers and de novo PUFA synthesis in colon cancer cells. Int J Mol Sci. 2019;20:832. doi: 10.3390/ijms20040832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jump DB, Tripathy S, Depner CM. Fatty acid-regulated transcription factors in the liver. Annu Rev Nutr. 2013;33:249–269. doi: 10.1146/annurev-nutr-071812-161139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tang C, Cho HP, Nakamura MT, Clarke SD. Regulation of human delta-6 desaturase gene transcription: identification of a functional direct repeat-1 element. J Lipid Res. 2003;44:686–695. doi: 10.1194/jlr.M200195-JLR200. [DOI] [PubMed] [Google Scholar]
- 81.Dong X, Tan P, Cai Z, Xu H, Li J, Ren W, Xu H, Zuo R, Zhou J, Mai K, Ai Q. Regulation of FADS2 transcription by SREBP-1 and PPAR-α influences LC-PUFA biosynthesis in fish. Sci Rep. 2017;7:40024. doi: 10.1038/srep40024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nakamura MT, Nara TY. Essential fatty acid synthesis and its regulation in mammals. Prostaglandins Leukot Essent Fatty Acids. 2003;68:145–150. doi: 10.1016/s0952-3278(02)00264-8. [DOI] [PubMed] [Google Scholar]
- 83.Brown NF, Stefanovic-Racic M, Sipula IJ, Perdomo G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 2007;56:1500–1507. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 84.Furuta E, Pai SK, Zhan R, Bandyopadhyay S, Watabe M, Mo YY, Hirota S, Hosobe S, Tsukada T, Miura K, Kamada S, Saito K, Iiizumi M, Liu W, Ericsson J, Watabe K. Fatty acid synthase gene is up-regulated by hypoxia via activation of Akt and sterol regulatory element binding protein-1. Cancer Res. 2008;68:1003–1011. doi: 10.1158/0008-5472.CAN-07-2489. [DOI] [PubMed] [Google Scholar]
- 85.Worgall TS, Sturley SL, Seo T, Osborne TF, Deckelbaum RJ. Polyunsaturated fatty acids decrease expression of promoters with sterol regulatory elements by decreasing levels of mature sterol regulatory element-binding protein. J Biol Chem. 1998;273:25537–25540. doi: 10.1074/jbc.273.40.25537. [DOI] [PubMed] [Google Scholar]
- 86.Jump DB. Fatty acid regulation of gene transcription. Crit Rev Clin Lab Sci. 2004;41:41–78. doi: 10.1080/10408360490278341. [DOI] [PubMed] [Google Scholar]
- 87.Triki M, Rinaldi G, Planque M, Broekaert D, Winkelkotte AM, Maier CR, Janaki Raman S, Vandekeere A, Van Elsen J, Orth MF, Grünewald TGP, Schulze A, Fendt SM. mTOR signaling and SREBP activity increase FADS2 expression and can activate Sapienate biosynthesis. Cell Rep. 2020;31:107806. doi: 10.1016/j.celrep.2020.107806. [DOI] [PMC free article] [PubMed] [Google Scholar]