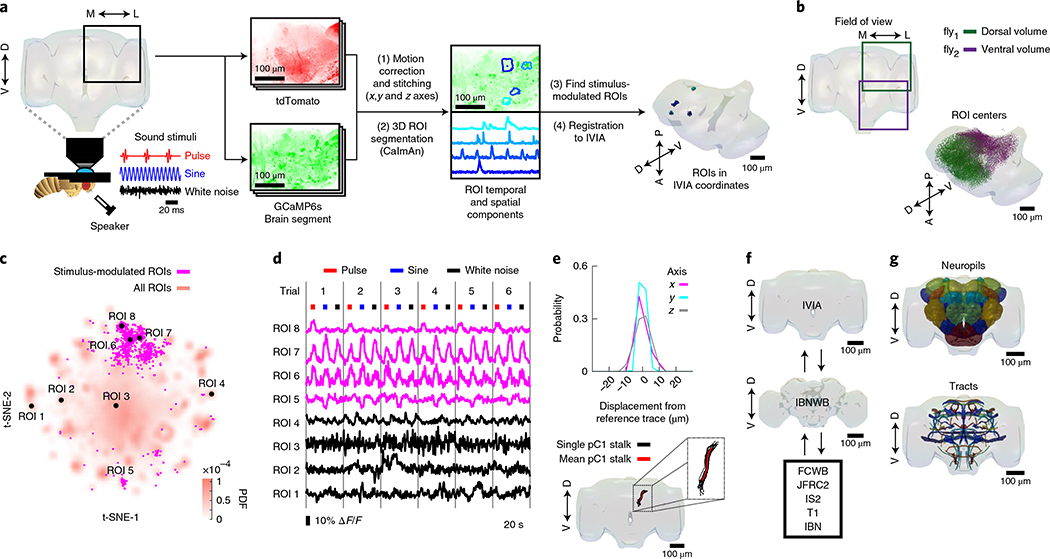
Fig. 1 |. A new pipeline for mapping sensory activity throughout the central brain.
a, Overview of data collection and processing pipeline (for more details see Extended Data Fig. 1a). Step (1): the tdTomato signal is used for motion correction of a volumetric time-series and to stitch serially imaged overlapping brain segments in the x, y and z axes. Step (2): 3D ROI segmentation (via CaImAn) is performed on GCaMP6s signals. Step (3): auditory ROIs are selected. Step (4): ROIs are mapped to the in vivo intersex atlas (IVIA) space. D, dorsal; L, lateral; M, medial; V, ventral. b, Top: an example of 11,225 segmented ROIs combined from two flies (dorsal (green) and ventral (purple) volumes from each fly). Bottom: ROI centers (dots) span the entire anterior–posterior (A–P) and D–V axes. These ROIs have not yet been sorted for those that are auditory (Extended Data Fig. 1d,e). c, 2D t-SNE embedding of activity from all ROIs in b. PDF, probability density function. ROIs modulated by auditory stimuli (1,118 out of 11,225 ROIs) are shown in magenta. d, ΔF/F from ROIs indicated in c. Magenta traces correspond to stimulus-modulated ROIs, while black traces are non-modulated ROIs. Time of individual auditory stimulus delivery is indicated (pulse, sine or band-limited white noise). e, Top: IVIA registration accuracy showing the per-axis jitter (x, y and z) between traced pC1 stalk values across flies relative to mean pC1 stalk values (see Extended Data Fig. 2a–c for more details). Bottom: 3D rendering of traced stalks of Dsx+ pC1 neurons (black traces, n = 20 brains from Dsx–GAL4/UAS–GCaMP6s flies) and mean pC1 stalk (red trace). f, Schematic of the bridging registration between the IVIA and the Insect Brain Nomenclature Whole Brain (IBNWB) atlas25. This bidirectional interface provides access to a network of brain atlases (FCWB, JFRC2, IS2, T1 and IBN) associated with different Drosophila neuroanatomy resources3. See Extended Data Fig. 2d–g for more details. g, Anatomical annotation of the IVIA. Neuropil and neurite tract segmentations from the IBN atlas were mapped to the IVIA (for a full list of neuropil and neurite tract names, see Supplementary Table 1). See also Supplementary Videos 1–4.