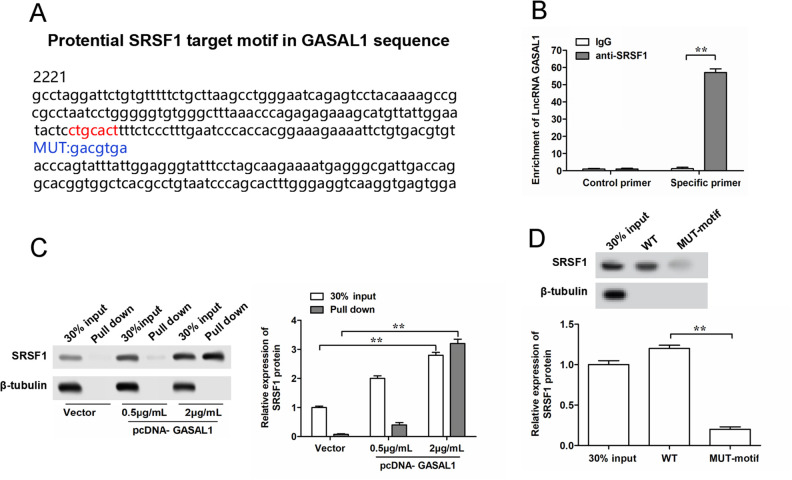
Figure 4.
Prediction and validation of binding between GASAL1 and SRSF1 protein in HTR-8/SVneo cells. (A) Schema of SRSF1 binding sites in the GASAL1 sequence. Red fonts represent the binding motifs, and blue fonts represent the mutated nucleotides in the binding motifs. (B) Binding of GASAL1 and SRSF1 was validated by SRSF1-antibody-based RNA-immunoprecipitation assay. (C) Binding of GASAL1 and SRSF1 was validated by GASAL1-probe-based RNA pull-down assay. (D) Binding of GASAL1 and SRSF1 was validated by RNA pull-down assay using WT or MUT biotinylated GASAL1 transcripts. The data were presented as the mean ± SEM, n = 3. Student’s t-test was used for comparisons between groups in this study.*P < 0.05 and **P < 0.01. GASAL1: lncRNA growth arrest associated lncRNA 1; lncRNA: long noncoding RNA; MUT: mutated; SRSF1: serine/arginine splicing factor 1; WT: wild type.