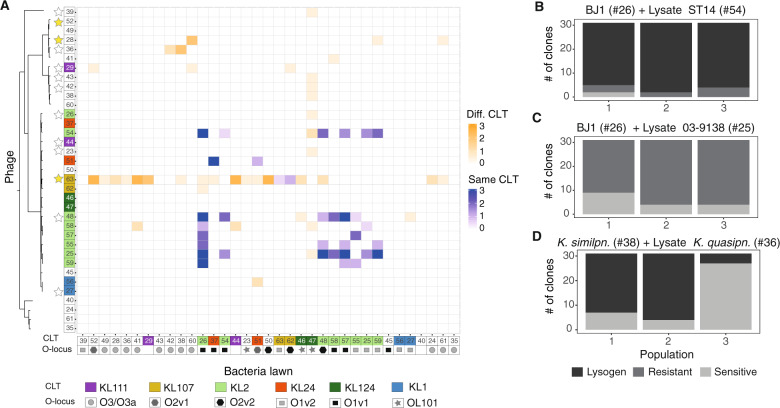
Fig. 4. Some Klebsiella encode viable prophages that can infect and lysogenize other strains.
a Infection matrix indicating the ability of induced and PEG-precipitated supernatants of all strains to form inhibition halos on lawns of all strains. This was repeated three times with three independently produced lysates. The sum of the three experiments is shown. The strains are ordered by phylogeny, and the colors on top of numbers indicate capsule locus types (CLT). Geometric figures along the x-axis indicate the LPS serotype (O-locus). Stars along the y-axis indicate that the genome codes for putative genes involved in colicin production but lacking a lysis gene (white) or has a full colicin operon (yellow). Shades of blue indicate infections between lysates produced by bacteria from the same CLT as the target bacteria, whereas shades of orange indicate cross-CLT infections. b–d Barplots indicate the number of resistant, sensitive, or lysogenized clones from each independent replicate population exposed 24 h to lysates from other strains. Each bar represents the 31 survivor clones from each of three independent experiments. Gray scale indicates whether: (i) the clones are still sensitive to the phage (light gray), (ii) they became resistant by becoming a lysogen (dark gray) as determined by cell death upon addition of MMC and PCR targeting prophage sequences, or (iii) they became resistant by other mechanisms.