FIGURE 3.
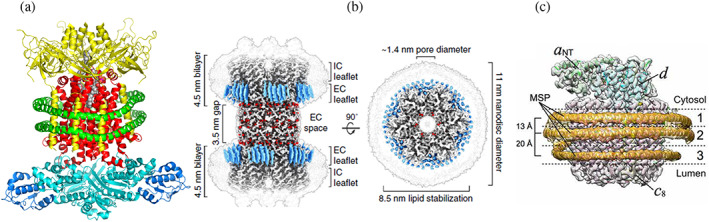
(a) Cryo‐electron microscopy (cryo‐EM) structure of the ABC transporter complex MlaFEDB. Hexamer MLAD (yellow), dimeric transport permease MLAE (red), ATP binding protein (cyan) and transporter‐binding protein MLAB (blue) are enclosed into MSP1D1 Nanodisc (two parallel helices shown in green). 112 (b) Cryo‐EM structure of connexin‐46/50 incorporated in two Nanodiscs (light gray contours). Well resolved lipids are shown in blue, pore with diameter 1.4 nm is shown as the top view on the right. Reproduced with permission from Flores et al. 113 (c) Cryo‐EM structure of the Vo proton channel of yeast vacuolar ATPAse. In this structure three molecules of MSP1E3D1 scaffold stabilize the protein complex by forming rings of different diameter around middle transmembrane part and around c‐ring. Reproduced with permission from Roh et al. 114 Open access article published under the CC BY license
