Abstract
SARS-CoV-2 membrane (M) protein performs a variety of critical functions in virus infection cycle. However, the expression and purification of membrane protein structure is difficult despite tremendous progress. In this study, the 3 D structure is modeled followed by intensive validation and molecular dynamics simulation. The lack of suitable homologous templates (>30% sequence identities) leads us to construct the membrane protein models using template-free modeling (de novo or ab initio) approach with Robetta and trRosetta servers. Comparing with other model structures, it is evident that trRosetta (TM-score: 0.64; TM region RMSD: 2 Å) can provide the best model than Robetta (TM-score: 0.61; TM region RMSD: 3.3 Å) and I-TASSER (TM-score: 0.45; TM region RMSD: 6.5 Å). 100 ns molecular dynamics simulations are performed on the model structures by incorporating membrane environment. Moreover, secondary structure elements and principal component analysis (PCA) have also been performed on MD simulation data. Finally, trRosetta model is utilized for interpretation and visualization of interacting residues during protein-protein interactions. The common interacting residues including Phe103, Arg107, Met109, Trp110, Arg131, and Glu135 in the C-terminal domain of M protein are identified in membrane-spike and membrane-nucleocapsid protein complexes. The active site residues are also predicted for potential drug and peptide binding. Overall, this study might be helpful to design drugs and peptides against the modeled membrane protein of SARS-CoV-2 to accelerate further investigation.
Communicated by Ramaswamy H. Sarma
Keywords: SARS-CoV-2 membrane protein, modeling approach, template-free modeling, model validation, molecular dynamics, principal component analysis, protein-protein interactions
Graphical Abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS CoV-2) with unpredictable and fast spreading nature has imposed the most devastating global impact in recent times. Thus, the pandemic has created a huge catastrophe for human life. Hence, targeting crucial viral proteins and exploring their structural features are the ongoing strategies to design effective vaccines or therapeutics. Three dimensional structure of a protein provides details insights from structure to function relationship which aids to the structure-based drug as well as vaccine design (Gromiha, 2010). The structural proteins’ interactions in SARS-CoV-2 may play critical role for the association of viral particles during viral replication and assembly (Li et al., 2020).
Membrane protein (M) is one of the important functional components that plays a significant role in maintaining virion size and shape. It assists to assemble all other structural proteins including spike (S), envelope (E), and nucleocapsid (N) and participates in the budding process (Neuman et al., 2011; Schoeman & Fielding, 2019). Coronaviruses form virus-like particle (VLP) via the interaction of M and E or M and N proteins, and the collective manifestation of M, N and E is mandatory for well-organized VLP production as well as its trafficking and release (Siu et al., 2008). In addition, M-S proteins’ interaction assist incorporation of S protein into virion. The M protein also collaborates with the S protein during the cell attachment and entry (Naskalska et al., 2019) and it seems that these crucial interaction may facilitate viral transmission. Moreover, viral M protein, like other viral proteins, exhibits self-association as well as interaction with other accessory and non-structural proteins. These protein-protein interactions may play a significant role in viral structural protein processing, modification, and trafficking for viral particle assembly and egress (Li et al., 2020). Thus, the critical network of SARS-CoV-2 M protein with its intra-viral proteins shapes the basis of targeting M protein as a target for structure-based drug design.
Even though the M protein is the indispensable part of its SARS-CoV-2 virion, the big bottleneck in designing structure-based drug is lack of its three-dimensional structure. So far there is no experimentally resolved structure as well as suitable template available for even homology modeling of M protein. Therefore, template-free modeling (de novo or ab initio) approach appears to be the most suitable to model M protein as no known structural homolog is available. Mostly, this approach applies physics based principles and energy terms to model proteins (Dhingra et al., 2020; Khor et al., 2015). The template-free modeling has exhibited drastic improvement of the accuracy for residue-residue contact distance prediction in the recent years. The actual prediction of inter-residue contacts and distances is a major intermediary step to predict protein three-dimensional (3D) structure from sequence (Hou et al., 2019).
In this study, we have utilized different modelling protocols including I-TASSER, Robetta, trRosetta, SWISS-MODEL to assess and compare the SARS-CoV-2 M protein model structures. Among theses protocols, I-TASSER applies multiple threading alignment approaches to build up the full-length protein model structures (Yang et al., 2015). While, Robetta protocol runs automated tools, and sequences submitted to the server are parsed into putative domains and structural models are assembled through either comparative modeling or de novo structure prediction approaches (Kim et al., 2004). trRosetta predicts inter-residue orientations and distances from co-evolutionary data applying deep knowledge, significantly improves protein structure prediction (Yang et al., 2020). Moreover, SWISS-MODEL looks for template against the template library (SMTL) applying BLAST and HHBlits. Then model is built using ProMod3 considering the target-template alignment. QMEAN scoring function assess the global and per-residue model quality to quantify modeling errors (Waterhouse et al., 2018). The predicted structures are verified via ERRAT, RAMPAGE, PROCHECK, ProSA-web (Wiederstein & Sippl, 2007), and QMEANBrane web servers (Studer et al., 2014; Zobayer & Hossain, 2018) as validation and quality assessment are the crucial task for three-dimensional structures (Pražnikar et al., 2019). In addition, all model structures are subjected to molecular dynamics simulation by incorporating the membrane environment.
2. Methods
2.1. Sequence analysis and domain identification
The SARS-CoV-2 membrane (M) protein sequence (YP_009724393.1) was retrieved from NCBI Reference Sequence (NC_045512.2) (Pruitt, Tatusova, & Maglott, 2007) and compared with SARS-CoV M protein sequence (UniProtKB-P59596) via BioEdit ClustalW application. The domain orientation of SARS-CoV-2 M protein was visualized based on UniProtKB- P0DTC5.
2.2. Physiochemical parameters and secondary structure analysis
To analyze physiochemical parameters ExPASy’s ProtParam (Gasteiger et al., 2005) tool was employed to calculate theoretical pI (Isoelectric point), instability index (II), aliphatic index (AI), grand average of hydropathicity for SARS-CoV-2 M protein. Furthermore, secondary structural properties of the protein were evaluated via self-optimized prediction method with alignment (SOPMA) (Dash et al., 2016).
2.3. Template search and alignment
BLAST (blastp) and SWISS-MODEL were searched to find the suitable template for SARS-CoV-2 M protein. The template library of SWISS-MODEL (SMTL) applied BLAST and HHBlits against the primary amino acid sequence in the library (Waterhouse et al., 2018). The 20 distant homologs were identified as probable template structures (Dilly et al., 2020).
2.4. Protein modeling and validation
The SARS-CoV-2 membrane (M) protein reference sequence (YP_009724393.1) was applied for template-free (de novo or ab initio) prediction of the 3 D structures employing Robetta and trRosetta servers. These model structures were also compared with the model generated by I-TASSER server. To assess quality of the predicted models, various validation servers including PROCHECK (Laskowski et al., 1993), RAMPAGE (Begum et al., 2019; Lovell et al., 2003), ERRAT (Colovos & Yeates, 1993), ProSA-web (Wiederstein & Sippl, 2007), and QMEANBrane (Studer et al., 2014) were used. Later, TM-align algorithm was also employed to identify the best model structures based on TM-score (Zhang & Skolnick, 2005).
2.5. Model refinement and energy minimization
Membrane proteins were refined and energy minimized by YASARA program (Land & Humble, 2018). For that purpose, the membrane was attached for all model structures. YASARA scanned for hydrophobic residues among the secondary structure elements of the protein that could be part of probable transmembrane region. YASARA displayed the suggested membrane embedding and built a membrane of the required size (69.2 Å × 7.3 Å) with the lipid composition of phosphatidyl-ethanolamine. An equilibration simulation was last for 250 ps. The membrane was stabilized to adapt the protein and maintain the right density during the equilibration phase.
2.6. Molecular dynamics (MD) simulation
YASARA Dynamics (Krieger et al., 2004) were used to perform the molecular dynamics simulation where AMBER14 force field (Dickson et al., 2014) was considered for all calculations. During the simulation, Berendsen thermostat process regulated the simulation temperature. The particle Mesh Ewald algorithm was involved for long-range electrostatic interactions. A periodic boundary condition was elected during the simulation of membrane embedded protein. The environment was equilibrated with 0.9% NaCl and water solvent, at 298 K temperature. The time step was about 1.25 fs to carry out 100 ns MD simulation and 1000 snapshots were collected at 100 ps time interval. After MD simulation, different data including root mean square deviation (RMSD), root mean square fluctuation (RMSF), solvent-accessible surface area (SASA), radius of gyration, total number of hydrogen bonds, helix, sheet, turn, and coil values were collected from MD simulations, according to previously published data analysis protocols (Ahmed, Islam, et al., 2020; M. J. Islam et al., 2019; R. Islam et al., 2020; Junaid et al., 2019; Khan et al., 2017; Shahinozzaman et al., 2020).
2.7. Principal component analysis (PCA)
MD simulation data were utilized for principal component analysis (PCA) to explore the structural and energy fluctuations among model M protein structures. The existent variability in the MD trajectory was observed by different multivariate energy factors in the low-dimensional space (De Jong, 1990; Wold et al., 1987). The centering and scaling were executed for data pre-processing (Ahmed, Mahtarin, et al., 2020; Chowdhury et al., 2020). In the analysis, final 90 ns MD trajectories were utilized to reveal the variations among the model structures. The PCA model is reflected by the following equation:
where X matrix expresses multivariate factors into the resultant of two new matrices, i.e. Tk and Pk; Tk represents matrix of scores which relates the samples; Pk, matrix of loadings correlates the variables, k is the number of factors presented in the model and E indicates the matrix of residuals. The exploration of trajectory was performed through R (Peng, 2015), RStudio (Rstudio Team, 2019) and internal codes. The PCA plots were originated using the R package ggplot2 (Wickham, 2009).
2.8. Protein-Protein interactions studies and active site determination of M protein
Protein-protein interactions and interacting residues of M protein with structural protein were investigated in the study. The molecular docking was performed among trRosetta model M protein and other full-length structural proteins (S, N, and E) from I-TASSER using PatchDock (Schneidman-Duhovny et al., 2005) then refined by FireDock (Mashiach et al., 2010). Further, proteins were also docked using ClusPro (Kozakov et al., 2017). The best poses were considered and visualized as the protein-protein complexes. The interacting residues among the protein complexes were exhibited by PDBsum’s interaction plots (Laskowski et al., 2018). The active site residues were also predicted as the probable drug binding site by CASTp web server (Wei Tian et al., 2018). We also retrieved protein-protein interactions and interactors for SARS-CoV-2 M protein (UniProtKB-P0DTC5) from IntAct Molecular Interaction Database (Aranda et al., 2010). Then the network was visualized using Cytoscape (version 3.8.0) (Cline et al., 2007).
3. Result
3.1. Analysis of sequence and domain region
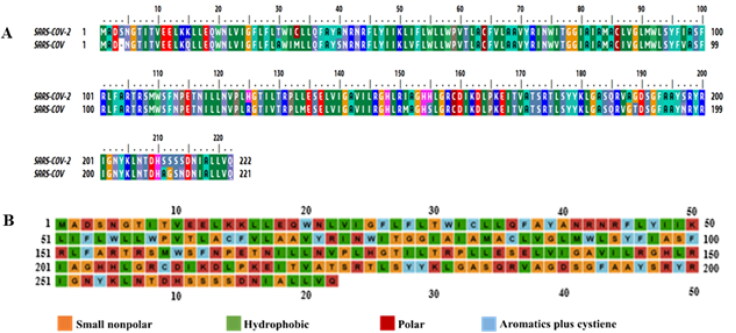
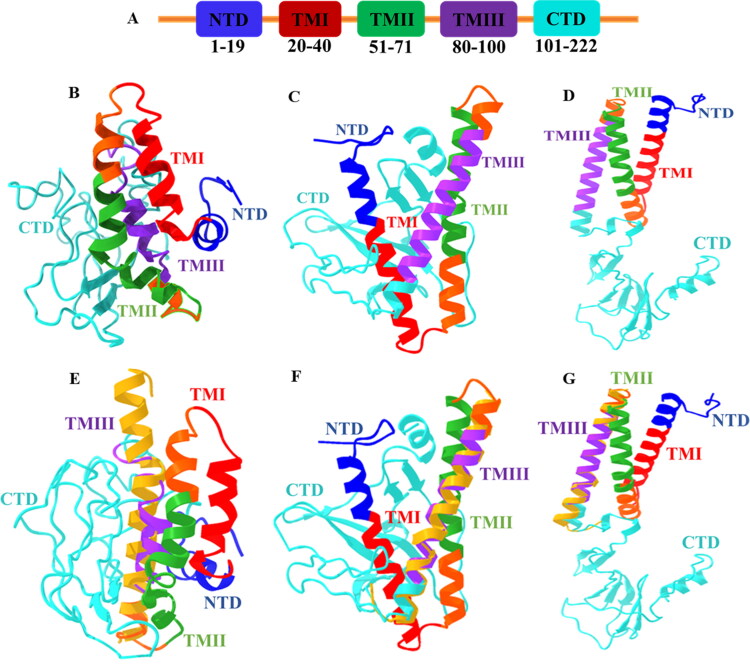
The membrane protein sequence (YP_009724393.1) of SARS-CoV-2 has shown, sequence identities are 90.5% and sequence similarities are about 96.40% compared with SARS-CoV M protein sequence (UniProtKB-P59596). The alignment is shown in Figure 1(a) applying BioEdit ClustalW application. It is observed that 20 mismatches and 1 gap in SARS-CoV-2 M protein comparing with SARS-CoV virus, possibly play a critical role in the virus infection cycle. Further, the domain regions of SARS-CoV-2 M protein (UniProtKB-P0DTC5) are demonstrated in Figure 2(a), where the N-terminal region covers (1–19) amino acids, three distinct transmembrane regions (TMI, TMII, TMIII) occupy regions in between (20–100) amino acids and C-terminal region resides within (101–222) amino acids.
Figure 1.
Sequence analysis. (a) Sequence alignment of SARS-CoV-2 (YP_009724393.1) and SARS-CoV (UniProtKB-P59596), (b) amino acid types.
Figure 2.
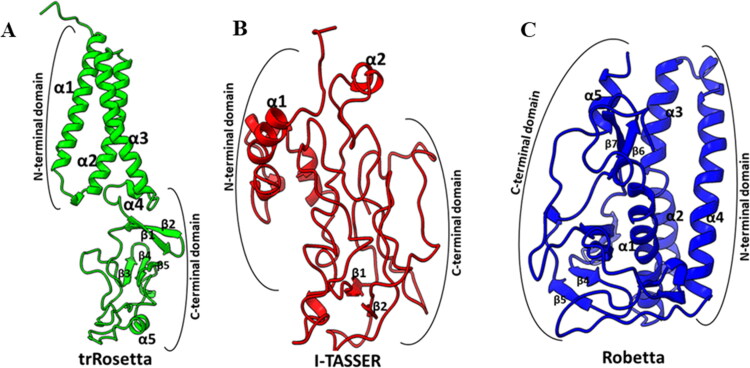
(a) Domain Organization of SARS-CoV-2 M protein. Each domain is labeled with corresponding amino acids, N-terminal region resides at exterior while C-terminal in interior of the virus particle and TM domain region is membrane embedded. Model (best) M protein structures (full-length) from (b) I-TASSER, (c) Robetta, and (d) trRosetta modeling servers. Structural comparison of model M proteins among (e) I-TASSER, (f) Robetta, and (g) trRosetta with model TM domain (golden) obtained from SWISS-MODEL.
3.2. Analysis of physiochemical parameters and secondary structure
The analysis of physicochemical parameters from ExPASy’s ProtParam has revealed that the M protein of SARS-CoV-2 has the isoelectric point 9.51, instability index 39.14, aliphatic index 120.86, grand average of hydrophobicity 0.446, and also has more positively charged residues (21) than negative-charged (13) amino acids. Moreover, the amino acids’ number and percentage of composition in the M protein sequence has been shown in (Table 1), where, the number and percentage (15.8%) of Leu residue are the highest among all residues. The annotated plots for amino acid types are visualized (Figure 1b). Moreover, the properties for secondary structure of the protein with the number of residues and percentages are also displayed via a self-optimized prediction method with alignment (SOPMA) (Table 2).
Table 1.
The amino acids’ number and composition of SARS-CoV-2 M protein from ExPASy’s ProtParam tool.
| Amino acid | Number | Composition |
|---|---|---|
| Ala (A) | 19 | 8.6% |
| Arg (R) | 14 | 6.3% |
| Asn (N) | 11 | 5.0% |
| Asp (D) | 6 | 2.7% |
| Cys (C) | 4 | 1.8% |
| Gln (Q) | 4 | 1.8% |
| Glu (E) | 7 | 3.2% |
| Gly (G) | 14 | 6.3% |
| His (H) | 5 | 2.3% |
| Ile (I) | 20 | 9.0% |
| Leu (L) | 35 | 15.8% |
| Lys (K) | 7 | 3.2% |
| Met (M) | 4 | 1.8% |
| Phe (F) | 11 | 5.0% |
| Pro (P) | 5 | 2.3% |
| Ser (S) | 15 | 6.8% |
| Thr (T) | 13 | 5.9% |
| Trp (W) | 7 | 3.2% |
| Tyr (Y) | 9 | 4.1% |
| Val (V) | 12 | 5.4% |
Table 2.
The secondary structure properties of the SARS-CoV-2 M protein by SOPMA.
| Structure | Number of residues | Percentage (%) |
|---|---|---|
| Alpha helix (Hh) | 77 | 34.68 |
| 310 helix (Gg) | 0 | 0.00 |
| Pi helix (Ii) | 0 | 0.00 |
| Beta bridge (Bb) | 0 | 0.00 |
| Extended strand (Ee) | 47 | 21.17 |
| Beta turn (Tt) | 15 | 6.76 |
| Bend region (Ss) | 0 | 0.00 |
| Random coil (Cc) | 83 | 37.39 |
| Ambiguous states (?) | 0 | 0.00 |
| Other states | 0 | 0.00 |
3.3. Template search and alignment
We have searched for suitable template through blastP suite against Protein Data Bank and SWISS-MODEL against the primary amino acid sequence in the library (SMTL), however, only top two templates (<30% sequence identity) for PDB ID:5CTG (Bidirectional sugar transporter SWEET2b) and PDB ID:6XDC (SARS-CoV-2 Protein 3a) are found. The 14.29% sequence identity has shown by bidirectional sugar transporter SWEET2b, which looks like almost transmembrane region (residues 74–109) and 15.63% sequence identity has revealed by SARS-CoV-2 Protein 3a, which looks like C-terminal region (residues 104–200) in Figure S1a and S1b. The alignment among the targets and models sequences has been displayed in Figure S2a and S2b.
3.4. Analysis of protein models and validation
Due to the unavailability of experimentally determined close homologs (>30% sequence identity), template-based modeling was not feasible for the membrane protein with the algorithms of MEDELLER, i-membrane, Memoir, and MODELLER. Therefore, SARS-CoV-2 M protein full-length model structure has been predicted through template-free modeling (de novo or ab initio) approach from Robetta and trRosetta servers. Each server has provided five model structures. In this study, the model structures are compared with the model generated by I-TASSER server (Figure 2b–d). Apparently, the models from Robetta and trRosetta are better than I-TASSER model considering the construction of their domain regions.
The accuracy of the models is determined by ERRAT, ProSA web, QMEANBrane, RAMPAGE, and PROCHECK Ramachandran plot. In Table 3, validation scores suggest that model 4 for Robetta and model 5 from trRosetta are the best models among the M protein structures. ERRAT server identifies incorrect regions of protein structures in random distributions of atoms, which can be differentiated from correct distributions. It has presented the scores in the range between 49.296–96.040% for the M protein models from I-TASSER, Robetta, and trRosetta (Figure S3). While RAMPAGE validates 3 D models according to geometry and deviation. It represents scores between 47.7–100% where 99.5 and 98.2% displayed by Robetta and trRosetta best models (Figure S4). Further, PROCHECK server has assessed the stereochemical quality of protein structures considering residue by residue and overall structural geometry. It exhibits results in the range from 35–96% where the best models from Robetta and trRosetta manifest 94 and 96% in the most favored regions (Figure S5). It is observed that I-TASSER has presented the lowest scores among the models of servers. However, the structural analysis from ProSA web has presented the z-score: −5.21, −4.2, and 4.11 for I-TASSER, Robetta, and trRosetta, respectively (Figure S6). The protein models are close to experimentally determined native conformers (NMR spectroscopy: dark blue). But it is difficult to identify a better model from this analysis as there is no previously resolved membrane protein structure for CoVs. Finally, membrane protein model assessment via QMEANBrane has revealed that tr-Rosetta model is properly embedded with membrane compared to Robetta and I-TASSER model proteins (Figure S7), thus tr-Rosetta model fulfills the criteria of membrane protein. This assessment has played an important role to decide about a better model of M protein. The local quality estimation by QMEANBrane usually exhibit scores in range [0,1] for good models. In that case, only trRosetta shows score 1 for proper membrane embedded scenario, whereas Robetta model exposes score nearby 1 but I-TASSER showed the lowest score among the models. Later, the full model structures are also aligned with the top models from SWISS-MODEL by TM-align server, where TM region (74–109) amino acid residues are aligned relatively better with trRosetta (TM-score: 0.64; RMSD 2 Å) model compared to Robetta (TM-score: 0.61; RMSD 3.3 Å) and I-TASSER models (TM-score: 0.45; RMSD 6.5 Å) (Figure 2e–g).
Table 3.
Model structures validation for SARS-CoV-2 M protein using different servers.
| Modeling server | Model | ERRAT (%) | RAMPAGE (%) | PROCHECK (%) | |
|---|---|---|---|---|---|
| I-TASSER | 1 | 49.296 | 47.7 | 35.8 | |
| Robetta | 1 | 90.291 | 98.2 | 90.5 | |
| 2 | 88.095 | 98.2 | 91 | ||
| 3 | 93.897 | 100 | 93 | ||
| 4 | 96.04 | 99.5 | 94 | ||
| 5 | 91.304 | 97.3 | 86.1 | ||
| trRosetta | 1 | 78.037 | 98.2 | 95.5 | |
| 2 | 76.571 | 96.4 | 94.5 | ||
| 3 | 78.977 | 96.4 | 94 | ||
| 4 | 78.409 | 98.2 | 95.5 | ||
| 5 | 80.571 | 98.2 | 96 | ||
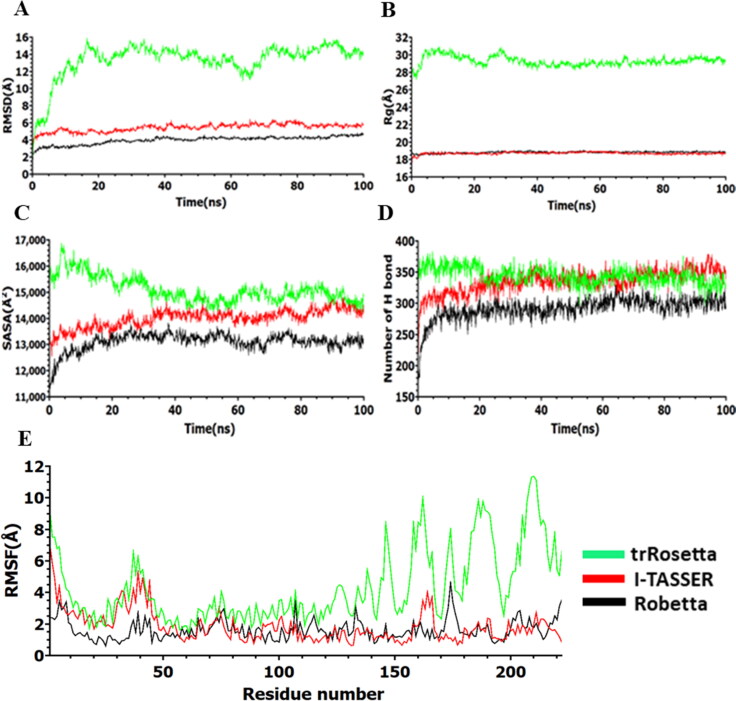
Figure 3.
Analysis of 100 ns simulation for model M protein structures. (a) Root mean square deviation values of C-α atom, (b) radius of gyration, (c) solvent accessible surface area, (d) number of hydrogen bonds, and (e) root mean square fluctuations during the simulation.
Figure 4.
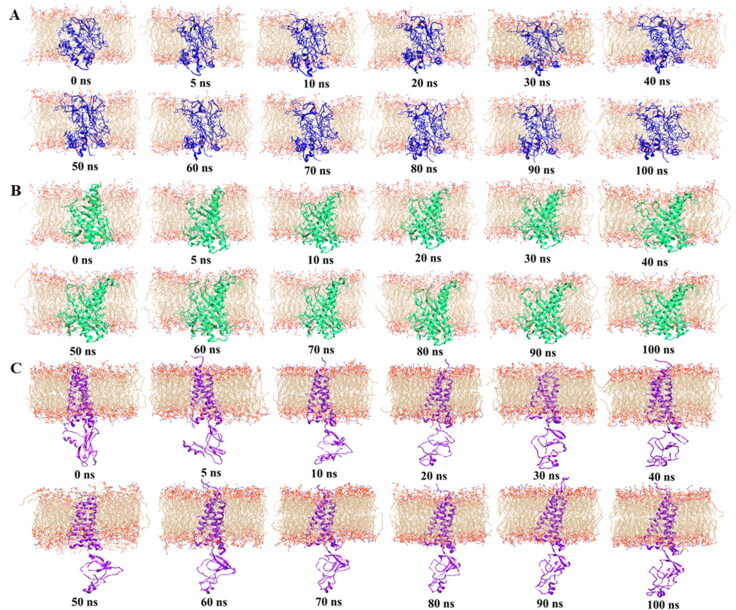
Membrane protein topology of SARS-CoV-2 over the 100 ns MD simulation for. (a) I-TASSER, (b) Robetta, and (c) trRosetta model structures.
Figure 5.
Representative secondary structure elements of model M proteins after MD simulation.
Figure 6.
PCA analysis on MD simulation. (a) The score plot represents three clusters for model M protein structures, where each dot indicates one time point. The clustering is attributable as: I-TASSER (orange), Robetta (blue), trRosetta (red). (b) Loading plot exhibits the energy and structural data from principal components analysis.
Figure 7.
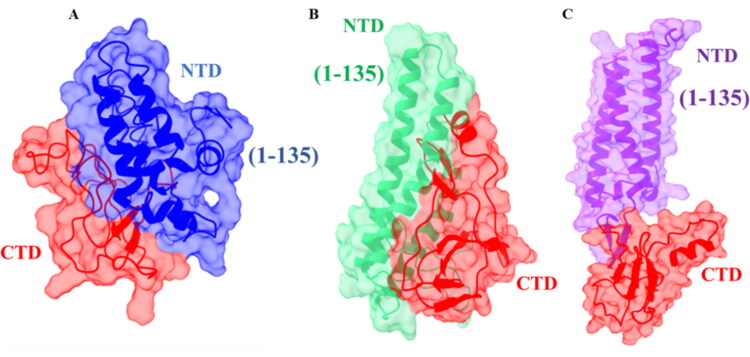
The important regions of M protein for interaction. (a) I-TASSER model (red and blue), (b) Robetta model (red and green), and (c) trRosetta model (red and purple) after 100 ns MD simulation. The 1–135 residues are important for interacting with M and S protein; where, I-TASSER model (blue), Robetta model (green) and trRosetta model (purple) indicate those crucial residues. The C-terminal cytosolic domain (residues 101–222) is important for interaction with envelope (E) and nucleocapsid (N) protein.
Currently, Rosetta is claimed to be the most successful template-free method in the CASP experiments (Lee et al., 2017; Kelm et al., 2014; Das & Baker, 2008). The deep learning-based prediction of inter-residue orientations, distances, and the improvement of a constrained optimization by Rosetta, can generate more accurate models for some template-free targets (Hou et al., 2019; Yang et al., 2020). Hence, considering all the perspectives, trRosetta model is the best model M protein structure in comparison with Robetta and I-TASSER model.
3.5. Molecular dynamics (MD) simulation analysis
We have performed 100 ns MD simulation to evaluate the probable conformational changes within each model structure from I-TASSER, Robetta, and trRosetta of M protein. RMSD values of α-carbon are investigated (Figure 3a). The higher average RMSD values of the Cα atoms are found for trRosetta model (∼13.34 Å), which is followed by I-TASSER model (∼7.68 Å) and Robetta model (∼3.98 Å) respectively. There is no higher and lower fluctuation observed for Robetta model and I-TASSER model over the simulation time, which suggests that these models are likely to be stable in an aqueous environment. On the other hand, trRosetta model shows significant deviation after 5 ns until 66 ns, where it begins to deviate largely. After that, fluctuation remains stable until the end of the simulation. High RMSD values are commonly observed in multidomain proteins where hinge motions produce relative movements of domains as rigid bodies (Lesk & Chothia, 1984; Monzon et al., 2017). When MD snapshots are analyzed, such changes are clearly observed in the C-terminal domain (Figure 4c). Moreover, the radius of gyration (Rg) of all trajectories is investigated to identify the degree of protein compactness. The Robetta and I-TASSER model has showed a similar pattern with lower Rg values compare to trRosetta model, indicating that compactness induced in the protein, as shown in Figure 3(b). Although trRosetta model exhibited distinct pattern with higher Rg values, the fluctuation shows a stable trend along the time. The SASA is calculated for all model structure and is depicted in Figure 3(c). The most prominent downtrends in the SASA have been observed in case of Robetta and I-TASSER model compare to trRosetta model, indicating that expansion of protein volume is lower than trRosetta model. The number of intramolecular hydrogen bonds is evaluated for deciphering the structural stability of the protein and plotted concerning the time (Figure 3d). The trRosetta (∼344) and I-TASSER (∼321) models have showed distinct pattern in the total number of H bonds over the simulation period while Robetta model showed the lowest number of H-bonds. RMSF calculation shows higher fluctuation for trRosetta model than I-TASSER and Robetta model which is presented in Figure 3(e). For trRosetta and I-TASSER model, N-terminal regions (1–24) and 25–55 residues of TM regions fluctuation patterns are almost overlapping, in contrast, these regions’ fluctuation patterns are distinct from Robetta model. The C-terminal region (104–222) shows significant fluctuation for trRosetta compared to other models. This result is consistent with RMSD result. However, the snapshots of model M protein structures (Figure 4) and their dynamic nature during the simulation period have been exhibited in movies (supplementary data).
3.6. Identification of secondary structure elements
The secondary structure elements are identified for MD simulated models of SARS-CoV-2 M protein, as shown in Figure 5. In trRosetta model, α-helices are showing in the positions 10–35, 40–70, 75–104, 109–112, 161–163, and 210–216 and β-sheets are in the positions 119–124, 127–132, 142–145, 148–151, and 154–156. On the other hand, I-TASSER model contains short length of α-helices in 10–19, and 99–105 and β-sheets in 143–145, and 193–195. In case of Robetta, the model shows five α-helices in the positions 12–36, 42–55, 59–71, 77–105, and 214-220 and nine β-sheets in the positions 122–123, 127–129, 140–145, 148–150, 155–158, 168–172, 176–180, 185–187, and 191–198, respectively. However, the dynamics of protein secondary structures have been observed for selected models over the simulation period as depicted in (Figure S8). In case of α-helix, tr-Rosetta has shown the highest average (42.34%) result as well as good stability compared with Robetta (40.86%) and I-TASSER (13.3%) models (Figure S8A), which might give overall stability to protein tertiary structure as well as more likely to be functional (Jochim & Arora, 2009). The higher average (22.57%) β-sheets are observed for Robetta model while irregular fluctuation observed in tr-Rosetta model (Figure S8b) in the C-terminal domain (Figure 5a). In case of turn and coil, I-TASSER model has exhibited a higher value compared to Robetta and tr-Rosetta models (Figure S8c–d).
Figure 8.
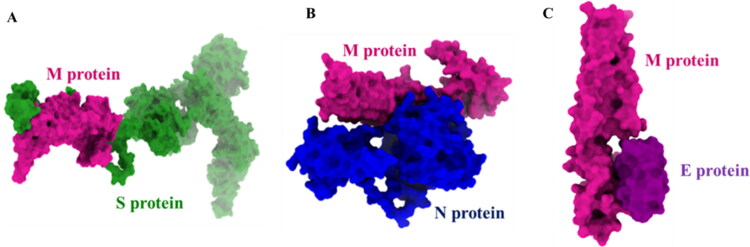
The snapshots of protein complexes after molecular docking. (a) M-S protein complexes, (b) M-N protein complexes, and (c) M-E protein complexes.
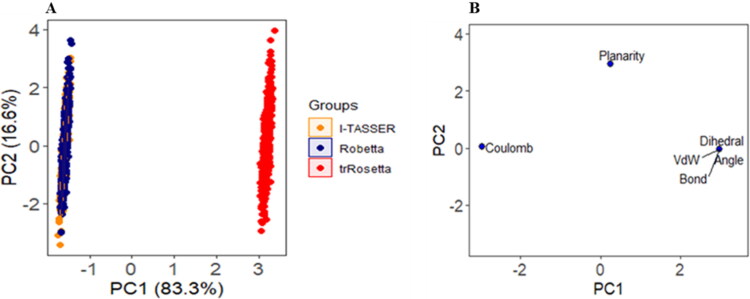
3.7. PCA analysis for constructed models of SARS-CoV-2 M protein
PCA analysis is used to realize structural and energy changes in models of SARS-CoV-2 M protein during MD simulation. Bond angles, bond distance, dihedral angles, planarity, van der Walls and electrostatic energies are included as variables. Here, PC1 and PC2 explain 99.9% of variance, where, PC1 exposes 83.3% and PC2 exposes 16.6% of variance. As shown in Figure 6(a), the score plot of PC1 and PC2 has demonstrated that a major rightward shifting found in trRosetta model compared to Robetta and I-TASSER models along PC1. This clustering pattern indicates majority of the variables including planarity, dihedral, angel, bond distance, and vdW energies have a higher influenced on the variance along PC1 (Figure 6b). On the other hand, Robetta and I-TASSER models are showing a similar pattern, its distribution clusters are at the farthest left, signifying the highest change in its coulomb energy profile.
3.8. Investigation of protein-protein interactions and active site residues
Previously, it has been observed that M and S protein of SARS-CoV co-expressed and the first 134 amino acids of M protein are crucial for their interaction (Voss et al., 2009). Hence, the corresponding interacting residues (1–135) in SARS-CoV-2 M are interpreted as N-terminal domain. Consequently, these 135 amino acids including three transmembrane domains are necessary to facilitate the accumulation of SARS-CoV M in the Golgi complex and to impose the recruitment of viral spike protein (S) to the sites of virus assembly and budding in the ERGIC (Satarker & Nampoothiri, 2020).
Comparing with SARS-CoV, the other structural proteins E and N of SARS-CoV-2 possibly interact with C-terminal domain (residues 100–222) of M protein (Fehr & Perlman, 2015; Schoeman & Fielding, 2019). This C-terminal domain has been recognized as a functional domain in M protein, which remains in the cytosol. As well as, this C-terminal polar tail within the endodomain interacts with S protein, which proposes that the large M endodomain (ME) possibly plays crucial roles in SARS-CoV assembly (Luo et al., 2006). Considering the location of C-terminal region, trRosetta model structure is more appropriate with its cytosolic domain while other models exhibit their unusual pattern. The highlighted domain regions of M protein which interacts with other structural proteins S, N, and E protein are shown in Figure 7(a)–(c).
In this study, interaction patterns of the proteins are visualized through docking approaches employing PatchDock, FireDock, and Cluspro2.0. Docking scores among trRosetta model M protein and other full-length structural proteins (S, N, and E) from I-TASSER are provided in Table S1. The expected best modes for protein complexes are pictured in Figure 8(a)–(c).
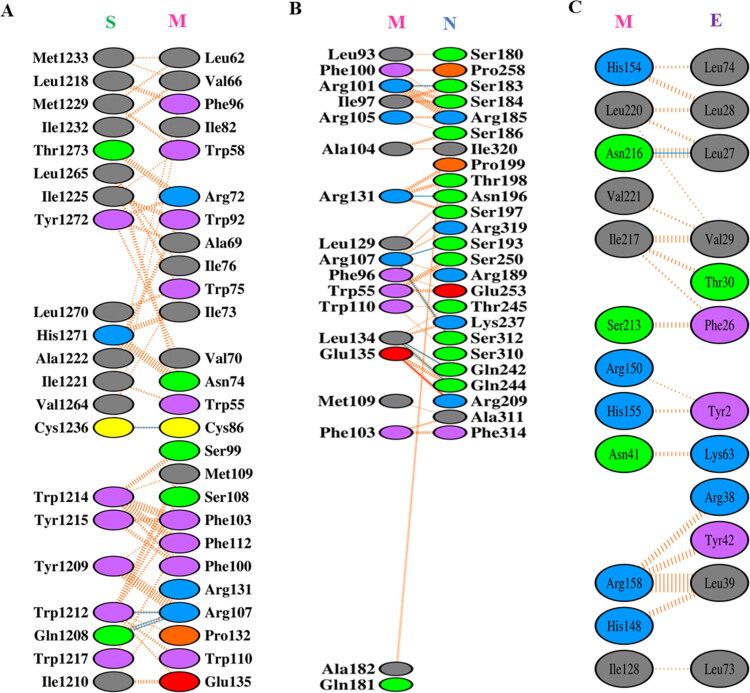
Later, the close view of interacting residues among the protein complexes is displayed through PDBsum’s interaction plots (Figure 9a–c). Among the complexes, M-S proteins interact through 4 H-bonds and 155 non-bonded contacts. The residues of M protein are remained within 1–135 while interacting with S protein. The M-N proteins’ interaction displays that 6 H-bonds, 181 non-bonded contacts, and a salt bridge are involved in the complex. In case of M-E interaction, 1 H-bond and 93 non-bonded contacts are responsible for proteins’ complex formation. The most common residual interactions of C-terminal domain of SARS-CoV-2 M are found in M-N and M-E complexes. The consequences of docking analysis are consistent with previous studies.
Figure 9.
Protein-protein interactions among M and other structural proteins S, N, and E after molecular docking. (a) Interaction between M and S proteins, (b) interaction between M and N proteins, and (c) interaction between M and E proteins.
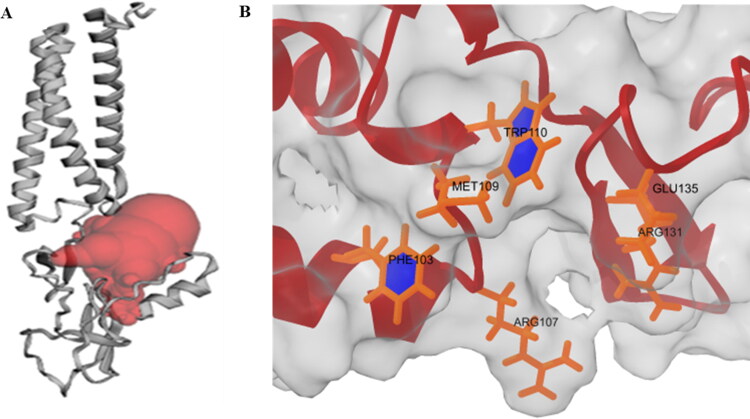
Moreover, in SARS-CoV, both E and N proteins are required to be co-expressed with M protein for the efficient assembly and release of VLPs. When these proteins are co-expressed, the native trimeric S glycoprotein is integrated into VLPs (Siu et al., 2008). Thus, the structural proteins’ role in VLP formation and infectivity is also predictable for SARS-CoV-2. Hence, we also aim to explore active site residues in trRosetta model M protein using CASTp web server. The prediction of active site residues will facilitate probable drug and peptide binding in the pocket. The residues and binding pocket are presented in Table S2 and Figure 10(a) accordingly. Besides, we have presented some common interacting residues, Phe103, Arg107, Met109, Trp110, Arg131, and Glu135 in C-terminal region of SARS-CoV-2 M protein. These residues are involved in the interaction with S and N structural proteins (Figure 10b).
Figure 10.
(a) Probable drug binding pocket (red) in trRosetta model M protein predicted by CASTp web server. (b) Commonly present residues in C-terminal region of M protein for interaction with S and N proteins.
Moreover, in recent times, it has been also observed that SARS-CoV-2 M protein and other structural proteins interact with accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF9b, and ORF10) as well as non-structural proteins (nsp2, nsp4, nsp5, nsp8, and nsp16). Therefore, these PPIs may play a significant role in viral structural protein processing, modification, and trafficking (Li et al., 2020). M protein also suppresses type I interferon (IFN) association by hindering the development of efficient TRAF3-involving complex (Siu et al., 2014).
Later, the 224 protein-protein interactions with M protein and 217 proteins have been retrieved from IntAct Molecular Interaction Database and interactions’ network is visualized through Cytoscape (version 3.8.0) in Figure S9. This network represents the abundant interaction of M protein with its intra viral and host interactome. These outcomes might expedite designing therapeutic strategies to disrupt the interaction among SARS-CoV-2 structural proteins as well as diverse interactome with other proteins.
4. Discussion
The modeling of accurate and reliable membrane proteins has been a great challenge since most of these protein structures have exhibited low sequence identities in PDB database (Berman et al., 2002). However, the computational modeling and prediction of three-dimensional structures of proteins holds the promise where experimental structures are not available (Schwede, 2013). In our study, we have employed template-free modeling strategy to model probable 3 D structure of (full-length) M protein and compared the model structure with the template-based models.
The SARS-CoV-2 M protein sequence was retrieved from the NCBI database and the details of protein id for further analysis are provided in the result section. In SARS-CoV-2 M protein sequence, 20 mismatches and 1 gap have been observed. These mutations in M protein could probably play a key role in viral infectivity and host cell interaction. The primary structure has been investigated and various parameters have been calculated using ExPasy ProtParam tool. The results suggested that SARS-CoV-2 M protein is basic with isoelectric point (pI) of 9.51. The amino acid composition showed the maximum presence of Leu (15.8%) and minimum presence of Cys, Gln, and Met (1.8%). Since, the crystal or cryo-EM structure of SARS-CoV-2 M protein does not solve yet, we have retrieved 3 D model for M protein from I-TASSER, Robetta, and trRosetta servers. The accuracy and quality of the structures are validated by employing various servers including ERRAT, RAMPAGE, PROCHECK, ProSA-web, and QMEANBrane. The ERRAT, PROCHECK, and RAMPAGE server assure about good quality of models suggesting most of residues in favoured regions. Moreover, ProSA-web calculates overall quality score of protein structure comparing with experimentally (X-ray, NMR) determined protein chains in PDB database. QMEANBrane evaluates the local quality of alpha-helical transmembrane protein models. It applies precisely trained potentials in a transmembrane protein model for three different segments (membrane, interface, and soluble).
From the validation analysis, it has been revealed that trRosetta model is the best model than others according to its proper orientation in the membrane environment. We have also compared the model structures with SWISS-MODEL top two models from PDB ID: 5CTG (Bidirectional sugar transporter SWEET2b) and 6XDC (SARS-CoV-2 Protein 3a). The model (74–109 residues) based on PDB ID: 5CTG (14.29% sequence identities) has displayed better alignment with trRosetta compared with the model from PDB ID: 6XDC.
Then, 100 ns MD simulation has been performed on trRosetta, I-TASSER and Robetta models. trRosetta model shows significant changes in RMSD with an average value of 13.34 Å than Robetta model (3.98 Å) and I-TASSER model (7.68 Å). Monzon et al. reported that in multidomain proteins with higher RMSD is very common because hinge motions produce relative movements of domains as rigid bodies (Monzon et al., 2017). However, RMSD is not a suitable measure for model quality assessment (Wallner & Elofsson, 2003) rather a comparatively good protein model with one bad region might render a very high RMSD (Moult et al., 2005). The Rg curves for trRosetta have showed to be much higher than Robetta and I-TASSER models. It is important to mention that all models are in a stable pattern which indicates a stable protein folding. The maximum Rg values designate loose packing of the protein conformation, which means structure that is more flexible (Dash et al., 2019). In case of SASA profile, trRosetta model shows a distinct pattern with a higher value of SASA over time, whereas Robetta and I-TASSER model presented lower SASA values. The decrease value of SASA indicates the shrunken nature of protein (Dash et al., 2019; Kamaraj & Purohit, 2013). We also observed a notable difference in H-bond pattern during the simulation period, whereas trRosetta model participates with a greater number of H-bonds, while I-TASSER and Robetta models showed lower participation in H-bonds interaction. Pace et al. reported that contribution of H-bonds to protein stability is strong (Pace et al., 2014). As can be seen in Figure 3(e), the plots of RMSF for trRosetta model show higher fluctuation with average RMSF value of 4.33 Å when compare to I-TASSER (2.12 Å) and Robetta (1.16 Å) models. We can notice in Figure 5(a) for trRosetta model, the C-terminal domain (104–222) contains most of the loop regions. However, the high fluctuation occurred in the loop regions (residue numbers; 146–149,160, 164–176,180–195, and 201–209) in the C-terminal domain for trRosetta. This is not unexpected because loop regions have lacked any definite geometry (Chowdhury et al., 2020). According to the RMSD, Rg, SASA, and RMSF plots results, the H-bond results of trRosetta, I-TASSER, and Robetta models have depicted that trRosetta conformation becomes more flexible and stable than I-TASSER and Robetta. This consequence is further supported by PCA analysis.
Further, the interacting residues and interactors for M protein have been explored. It has been interpreted that the first 135 amino acids are crucial for M and S protein interactions. This region has been adequate to mediate the accumulation of M in the Golgi complex. Thus, imposing the recruitment of the viral S protein to the directions of virus assembly and budding in the ERGIC (Voss et al., 2009). Besides, the C-terminal region is the functional domain for interacting with E and N structural proteins. Moreover, M-N interaction stabilizes the nucleocapsid (N protein-RNA complex), and also the internal core of virions, eventually, promotes completion of viral assembly (Escors et al., 2001; Fehr & Perlman, 2015). This indicates C-terminal domain of the M protein structure must be cytosolic. The molecular docking study for M protein with other structural proteins (S, E, and N) supports our interpretation from previous studies. Moreover, the common interacting residues Phe103, Arg107, Met109, Trp110, Arg131, and Glu135 of M protein in C-terminal domain are identified and visualized for interaction with S and N. The active site prediction by CASTp has been supposed to be crucial for targeting the SARS-CoV-2 M protein. We have also visualized the network of interactions for M protein (UniProtKB-P0DTC5) with other cellular proteins using Cytoscape (version 3.8.0). The interactions of ORF3a and M protein have also displayed the structural functions for SARS-CoV virus (Huang et al., 2006), finding relevancy with SARS-CoV-2 in our study. Consequently, the insights into atomic detail of the three-dimensional structures for proteins are crucial for a better understanding of biological processes. Only the accurate structures can be intensely used to sort out biological queries (Pražnikar et al., 2019).
In this study, considering structural pattern for TM region and cytosolic C-terminal region, trRosetta (model 5; TM score: 0.64; TM region RMSD: 2 Å) has provided relatively better model than Robetta (model 4; TM score: 0.61; TM region RMSD: 3.3 Å) and I-TASSER (TM score: 0.45; TM region RMSD: 6.5 Å). Recently, a research group reported that trRosetta and AlphaFold M protein models have displayed almost similar patterns of structures. Contrary to these models, I-TASSER model from Zhang group has manifested poor local geometries, poor side-chain conformations, bad backbone dihedral angles, and numerous atomic clashes generally suggested poor stereochemistry (Heo & Feig, 2020).
Furthermore, the models with good quality, particularly in the TM region which can be produced even in the target-template sequence identity 20 − 40% region (Nikolaev et al., 2018). Another study has obtained more accurate alignments for proteins with low sequence identities to their templates, can be achieved using structure-based profile alignment methods. This one has correlations with our study in which, modeling of structure is at least acceptable to membrane proteins where models exhibit RMSD-Cα values to the native of 2 Å or less in the transmembrane regions (Forrest et al., 2006).
However, the common error sources are alignment errors, backbone distortions, misplaced side chains, or picking a template of incorrect fold with low sequence identity and high structural divergent model (Al-Khayyat & Al-Dabbagh, 2016). Melo et al. has mentioned that typical errors in the model are either in or close to regions that join secondary structure central parts and are of high energy (Melo et al., 1997). Conversely, the models with a slightly higher (worse) RMSD but nearly correct overall fold may be used for prediction of function from their global fold, (Kihara & Skolnick, 2004) categorization of local functional sites (Weidong Tian et al., 2004; Li et al., 2008), or analyzing low-resolution structure (Shin et al., 2017).
The membrane protein of SARS-CoV-2 is one of the vital proteins, advances in the 3 D structure determination might speed up the drug discovery process. Now, computational prediction of the protein structure can play a central role in its structural elucidation (Muhammed & Aki-Yalcin, 2019). We have explored reliable and extensively employed computational methods to explore and evaluate the probable M protein structure for SARS-CoV-2 for further application.
5. Conclusion
This study elucidates the structural and dynamic features of SARS-CoV-2 M protein. To explore the biological consequences, in-depth realization of structural phenomenon is indispensable. In this study, we have employed in silico approaches for modeling of M protein. The models are extensively evaluated through ERRAT, RAMPAGE, PROCHECK, ProSA-web, and QMEANBrane servers. The best models from Robetta and trRosetta are further considered for MD simulation analysis comparing with I-TASSER server model. Our results disclose that M protein model generated from trRosetta is comparatively better than the models generated from Robetta and I-TASSER servers. Moreover, the utility of trRosetta model structure is interpreted through visualization of interacting residues during protein-protein interactions. This study provides details structural and dynamics insights of SARS-CoV-2 M protein which may help designing potent and selective inhibitors targeting the membrane protein.
Supplementary Material
Acknowledgements
We are grateful to our donors (http://grc-bd.org/donate/) who supported to build a computational platform. The authors like to acknowledge The World Academy of Science (TWAS) to purchase the High-Performance Computers for molecular dynamics simulation. We also like to give special thanks to Mst. Noorjahan Begum, Research Investigator, Virology Laboratory, International Centre for Diarrhoeal Disease Research, Mohakhali, Dhaka, Bangladesh; Md. Waseque Mia, Assistant Professor, Dept. of Biochemistry and Molecular Biology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh; Md Nayeem Hossain, Sayeda Samina Ahmed, Rajib Islam, Md. Rimon Parvez, Division of Infectious Diseases and Division of Computer Aided Drug Design, The Red-Green Research Centre, Dhaka, Bangladesh for their cordial support and valuable suggestions.
Disclosure statement
The authors declare no competing financial interest.
References
- Ahmed, S., Islam, N., Shahinozzaman, M., Fakayode, S. O., Afrin, N., & Halim, M. A. (2020). Virtual screening, molecular dynamics, density functional theory and quantitative structure activity relationship studies to design peroxisome proliferator-activated receptor-γ agonists as anti-diabetic drugs. Journal of Biomolecular Structure and Dynamics, 1–15. 10.1080/07391102.2020.1714482 [DOI] [PubMed] [Google Scholar]
- Ahmed, S., Mahtarin, R., Ahmed, S. S., Akter, S., Islam, M.S., Mamun, A. Al, Islam, R., Hossain, M. N., Ali, M. A., Sultana, M. U. C., Parves, M. R., Ullah, M. O., Halim, M. A. (2020). Investigating the binding anity, interaction, and structure-activity-relationship of 76 prescription antiviral drugs targeting RdRp and Mpro of SARS-CoV-2. J. Biomol. Struct. Dyn, 1–16. 10.1080/07391102.2020.1796804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khayyat, M. Z. S., & Al-Dabbagh, A. G. A. (2016). In silico prediction and docking of tertiary structure of LuxI, an inducer synthase of Vibrio fischeri. Reports of Biochemistry & Molecular Biology, 4(2), 66–75. [PMC free article] [PubMed] [Google Scholar]
- Aranda, B., Achuthan, P., Alam-Faruque, Y., Armean, I., Bridge, A., Derow, C., Feuermann, M., Ghanbarian, A. T., Kerrien, S., Khadake, J., Kerssemakers, J., Leroy, C., Menden, M., Michaut, M., Montecchi-Palazzi, L., Neuhauser, S. N., Orchard, S., Perreau, V., Roechert, B., … Hermjakob, H. (2010). The IntAct molecular interaction database in 2010. Nucleic Acids Research, 38(suppl_1), D525–531. 10.1093/nar/gkp878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begum, M. N., Islam, M. T., Hossain, S. R., Bhuyan, G. S., Halim, M. A., Shahriar, I., Sarker, S. K., Haque, S., Konika, T. K., Islam, M. S., Rahat, A., Qadri, S. K., Sultana, R., Begum, S., Sultana, S., Saha, N., Hasan, M., Hasanat, M. A., Banu, H., … Mannoor, K. (2019). Mutation spectrum in TPO gene of Bangladeshi patients with thyroid dyshormonogenesis and analysis of the effects of different mutations on the structural features and functions of TPO protein through in silico approach. BioMed Research International, 2019, 9218903. 10.1155/2019/9218903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman, H. M., Bluhm, W. F., Philip, E., Marvin, J., Weissig, H., & John, D. (2002). Research papers: The Protein Data Bank. Acta Crystallographica Section D 58, 899–907. [DOI] [PubMed] [Google Scholar]
- Chowdhury, S. M., Talukder, S. A., Khan, A. M., Afrin, N., Ali, M. A., Islam, R., Parves, R., Al Mamun, A., Sufian, M. A., Hossain, M. N., Hossain, M. A., & Halim, M. A. (2020). Antiviral peptides as promising therapeutics against SARS-CoV-2. The Journal of Physical Chemistry B, 124(44), 9785–9792. 10.1021/acs.jpcb.0c05621 [DOI] [PubMed] [Google Scholar]
- Cline, M. S., Smoot, M., Cerami, E., Kuchinsky, A., Landys, N., Workman, C., Christmas, R., Avila-Campilo, I., Creech, M., Gross, B., Hanspers, K., Isserlin, R., Kelley, R., Killcoyne, S., Lotia, S., Maere, S., Morris, J., Ono, K., Pavlovic, V., … Bader, G. D. (2007). Integration of biological networks and gene expression data using cytoscape. Nature Protocols, 2(10), 2366–2382. 10.1038/nprot.2007.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos, C., & Yeates, T. O. (1993). Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Science : A Publication of the Protein Society, 2(9), 1511–1519. 10.1002/pro.5560020916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, R., & Baker, D. (2008). Macromolecular modeling with rosetta. Annu. Rev. Biochem., 77, 363–382. [DOI] [PubMed] [Google Scholar]
- Dash, R., Ali, M. C., Dash, N., Azad, M. A. K., Zahid Hosen, S. M., Hannan, M. A., & Moon, I. S. (2019). Structural and dynamic characterizations highlight the deleterious role of SULT1A1 R213H polymorphism in substrate binding. International Journal of Molecular Sciences, 20(24), 6256. 10.3390/ijms20246256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash, R., Hosen, S. M. Z., Sultana, T., Junaid, M., Majumder, M., Ishat, I. A., & Uddin, M. M. N. (2016). Computational analysis and binding site identification of type III secretion system ATPase from Pseudomonas aeruginosa. Interdisciplinary Sciences, Computational Life Sciences, 8(4), 403–411. 10.1007/s12539-015-0121-z [DOI] [PubMed] [Google Scholar]
- De Jong, S. (1990). Multivariate calibration, H. Martens and T. Naes, Wiley. (1989). ISBN 0 471 90979 3. Price: £75.00, US$138.00. No. of pages: 504. Journal of Chemometrics, 4(6), 441–441. 10.1002/cem.1180040607 [DOI] [Google Scholar]
- Dhingra, S., Sowdhamini, R., Cadet, F., & Offmann, B. (2020). A glance into the evolution of template-free protein structure prediction methodologies. Biochimie, 175, 85–92. 10.1016/j.biochi.2020.04.026 [DOI] [PubMed] [Google Scholar]
- Dickson, C. J., Madej, B. D., Skjevik, Å. A., Betz, R. M., Teigen, K., Gould, I. R., & Walker, R. C. (2014). Lipid14: The Amber lipid force field. Journal of Chemical Theory and Computation, 10(2), 865–879. 10.1021/ct4010307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilly, S., Garnier, M., Solé, M., Bailly, R., Taib, N., & Bestel, I. (2020). In silico identification of a key residue for substrate recognition of the riboflavin membrane transporter RFVT3. Journal of Chemical Information and Modeling, 60(3), 1368–1375. 10.1021/acs.jcim.9b01020 [DOI] [PubMed] [Google Scholar]
- Escors, D., Ortego, J., Laude, H., & Enjuanes, L. (2001). The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. Journal of virology, 75(3), 1312–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An overview of their replication and pathogenesis. Coronaviruses: Methods and Protocols, 1282, 1–23. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, L. R., Tang, C. L., & Honig, B. (2006). On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophysical Journal, 91(2), 508–517. 10.1529/biophysj.106.082313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D., & Bairoch, A. (2005). The proteomics protocols handbook (pp. 571–608). Springer. 10.1385/1592598900 [DOI] [Google Scholar]
- Gromiha, M. M. (2010). Protein bioinformatics from sequence to function. Books Free. [Google Scholar]
- Heo, L., & Feig, M. (2020). Modeling of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins by machine learning and physics-based refinement. [DOI] [PMC free article] [PubMed]
- Hou, J., Wu, T., Cao, R., & Cheng, J. (2019). Protein tertiary structure modeling driven by deep learning and contact distance prediction in CASP13. Proteins, 87(12), 1165–1178. 10.1002/prot.25697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C., Narayanan, K., Ito, N., Peters, C. J., & Makino, S. (2006). Severe acute respiratory syndrome coronavirus 3a protein is released in membranous structures from 3a protein-expressing cells and infected cells. Journal of Virology, 80(1), 210–217. 10.1128/JVI.80.1.210-217.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, M. J., Khan, A. M., Parves, M. R., Hossain, M. N., & Halim, M. A. (2019). Prediction of deleterious non-synonymous SNPs of human STK11 gene by combining algorithms, molecular docking, and molecular dynamics simulation. Scientific Reports, 9(1), 16426. 10.1038/s41598-019-52308-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam, R., Parves, R., Paul, A. S., Uddin, N., Rahman, M. S., Mamun, A. A., … Halim, M. A. (2020). A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. Journal of Biomolecular Structure and Dynamics, 1–20. 10.1080/07391102.2020.1761883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochim, A. L., & Arora, P. S. (2009). Assessment of helical interfaces in protein-protein interactions. Molecular Biosystems, 5(9), 924–926. 10.1039/b903202a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junaid, M., Islam, N., Hossain, M. K., Ullah, M. O., & Halim, M. A. (2019). Metal based donepezil analogues designed to inhibit human acetylcholinesterase for Alzheimer’s disease. PLOS One, 14(2), e0211935. 10.1371/journal.pone.0211935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamaraj, B., & Purohit, R. (2013). In silico screening and molecular dynamics simulation of disease-associated nsSNP in TYRP1 gene and its structural consequences in OCA3. BioMed Research International, 2013, 697051. 10.1155/2013/697051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelm, S., Choi, Y., & Deane, C. M. (2014). Protein Modeling and Structural Prediction. In Springer Handbook of Bio-/Neuroinformatics (pp. 171–182). Berlin, Heidelberg: Springer. [Google Scholar]
- Khan, A. M., Shawon, J., & Halim, M. A. (2017). Multiple receptor conformers based molecular docking study of fluorine enhanced ethionamide with mycobacterium enoyl ACP reductase (InhA). Journal of Molecular Graphics and Modelling, 77, 386–398. 10.1016/j.jmgm.2017.09.010 [DOI] [PubMed] [Google Scholar]
- Khor, B. Y., Tye, G. J., Lim, T. S., & Choong, Y. S. (2015). General overview on structure prediction of twilight-zone proteins. Theoretical Biology and Medical Modelling, 12(1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara, D., & Skolnick, J. (2004). Microbial genomes have over 72% structure assignment by the threading algorithm PROSPECTOR_Q. Proteins, 55(2), 464–473. 10.1002/prot.20044 [DOI] [PubMed] [Google Scholar]
- Kim, D. E., Chivian, D., & Baker, D. (2004). Protein structure prediction and analysis using the Robetta server. Nucleic Acids Research, 32(Web Server issue), W526–W531. 10.1093/nar/gkh468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov, D., Hall, D. R., Xia, B., Porter, K. A., Padhorny, D., Yueh, C., Beglov, D., & Vajda, S. (2017). The ClusPro web server for protein–protein docking. Nature Protocols, 12(2), 255–278. 10.1038/nprot.2016.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger, E., Darden, T., Nabuurs, S. B., Finkelstein, A., & Vriend, G. (2004). Making optimal use of empirical energy functions: Force-field parameterization in crystal space. Proteins, 57(4), 678–683. 10.1002/prot.20251 [DOI] [PubMed] [Google Scholar]
- Land, H., & , Humble, M. S. (2018). YASARA: A Tool to Obtain Structural Guidance in Biocatalytic Investigations. Methods in Molecular Biology (Clifton, N.J.), 1685, 43–67. 10.1007/978-1-4939-7366-8_429086303 [DOI] [PubMed] [Google Scholar]
- Laskowski, R. A., Jabłońska, J., Pravda, L., Vařeková, R. S., & Thornton, J. M. (2018). PDBsum: Structural summaries of PDB entries. Protein Science: A Publication of the Protein Society, 27(1), 129–134. 10.1002/pro.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S., & Thornton, J. M. (1993). PROCHECK: A program to check the stereochemical quality of protein structures. Journal of Applied Crystallography, 26(2), 283–291. 10.1107/S0021889892009944 [DOI] [Google Scholar]
- Lee, J., Freddolino, P. L., & Zhang, Y. (2017). Ab initio protein structure prediction. 10.1007/978-94-024-1069-3 [DOI]
- Lesk, A. M., & Chothia, C. (1984). Mechanisms of domain closure in proteins. Journal of Molecular Biology, 174(1), 175–191. 10.1016/0022-2836(84)90371-1 [DOI] [PubMed] [Google Scholar]
- Li, B., Turuvekere, S., Agrawal, M., La, D., Ramani, K., & Kihara, D. (2008). Characterization of local geometry of protein surfaces with the visibility criterion. Proteins: Structure, Function, and Bioinformatics, 71(2), 670–683. [DOI] [PubMed] [Google Scholar]
- Li, J., Guo, M., Tian, X., Wang, X., Yang, X., Wu, P., Liu, C., Xiao, Z., Qu, Y., Yin, Y., Wang, C., Zhang, Y., Zhu, Z., Liu, Z., Peng, C., Zhu, T., & Liang, Q. (2020). Virus-host interactome and proteomic survey of PBMCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med 1, 1–15. 10.1016/j.medj.2020.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell, S. C., Davis, I. W., Adrendall, W. B., de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S., & Richardson, D. C. (2003). Structure validation by C alpha geometry: Phi, psi and C beta deviation. Proteins-Structure Function and Genetics, 50(3), 437–450. 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Luo, H., Wu, D., Shen, C., Chen, K., Shen, X., & Jiang, H. (2006). Severe acute respiratory syndrome coronavirus membrane protein interacts with nucleocapsid protein mostly through their carboxyl termini by electrostatic attraction. The International Journal of Biochemistry & Cell Biology, 38(4), 589–599. 10.1016/j.biocel.2005.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiach, E., Nussinov, R., & Wolfson, H. J. (2010). FiberDock: A web server for flexible induced-fit backbone refinement in molecular docking. Nucleic Acids Research, 38(Web Server), W457–461. 10.1093/nar/gkq373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo, F., Devos, D., Depiereux, E., Feytmans, E. (1997). ANOLEA: A www server to assess protein structures. Proceedings of International Conference on Intelligent Systems for Molecular Biology; ISMB. International Conference on Intelligent Systems for Molecular Biology, 5, pp. 187–190. [PubMed] [Google Scholar]
- Monzon, A. M., Zea, D. J., Fornasari, M. S., Saldaño, T. E., Fernandez-Alberti, S., Tosatto, S. C. E., & Parisi, G. (2017). Conformational diversity analysis reveals three functional mechanisms in proteins. PLoS Computational Biology, 13(2), e1005398. 10.1371/journal.pcbi.1005398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moult, J., Fidelis, K., Rost, B., Hubbard, T., & Tramontano, A. (2005). Critical assessment of methods of protein structure prediction (CASP) - Round 6. Proteins: Structure, Function, and Bioinformatics, 61(S7), 3–7. 10.1002/prot.20716 [DOI] [PubMed] [Google Scholar]
- Muhammed, M. T., & Aki-Yalcin, E. (2019). Homology modeling in drug discovery: Overview, current applications, and future perspectives. Chemical Biology & Drug Design, 93(1), 12–20. 10.1111/cbdd.13388 [DOI] [PubMed] [Google Scholar]
- Naskalska, A., Dabrowska, A., Szczepanski, A., Milewska, A., Jasik, K. P., & Pyrc, K. (2019). Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor. Journal of Virology, 93(19), 355–374. 10.1128/JVI.00355-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman, B. W., , Kiss, G., , Kunding, A. H., , Bhella, D., , Baksh, M. F., , Connelly, S., , Droese, B., , Klaus, J. P., , Makino, S., , Sawicki, S. G., , Siddell, S. G., , Stamou, D. G., , Wilson, I. A., , Kuhn, P., & , Buchmeier, M. J. (2011). A structural analysis of M protein in coronavirus assembly and morphology. Journal of Structural Biology, 174(1), 11–22. https://doi.org/ 10.1016/j.jsb.2010.11.02121130884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev, D. M., Shtyrov, A. A., Panov, M. S., Jamal, A., Chakchir, O. B., Kochemirovsky, V. A., … Ryazantsev, M. N. (2018). A comparative study of modern homology modeling algorithms for rhodopsin structure prediction. ACS Omega, 3(7), 7555–7566. 10.1021/acsomega.8b00721 [DOI] [PMC free article] [PubMed]
- Pace, C. N., Fu, H., Lee Fryar, K., Landua, J., Trevino, S. R., Schell, D., Thurlkill, R. L., Imura, S., Scholtz, J. M., Gajiwala, K., Sevcik, J., Urbanikova, L., Myers, J. K., Takano, K., Hebert, E. J., Shirley, B. A., & Grimsley, G. R. (2014). Contribution of hydrogen bonds to protein stability. Protein Science: A Publication of the Protein Society, 23(5), 652–661. 10.1002/pro.2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, R. D. (2015). R programming for data science, 109-116. [Google Scholar]
- Pražnikar, J., Tomić, M., & Turk, D. (2019). Validation and quality assessment of macromolecular structures using complex network analysis. Scientific Reports, 9(1), 1–11. 10.1038/s41598-019-38658-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt, K. D., Tatusova, T., & Maglott, D. R. (2007). NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic acids research, 35(suppl_1), D61-D65. 10.1093/nar/gkl842 [DOI] [PMC free article] [PubMed]
- Rstudio Team. (2019). RStudio: Integrated development for R. RStudio, Inc. 10.1007/978-3-642-20966-6 [DOI] [Google Scholar]
- Satarker, S., & Nampoothiri, M. (2020). Structural Proteins in Severe Acute Respiratory Syndrome Coronavirus-2. Archives of Medical Research, 51(6), 482–491. 10.1016/j.arcmed.2020.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneidman-Duhovny, D., Inbar, Y., Nussinov, R., & Wolfson, H. J. (2005). PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Research, 33(Web Server issue), W363–W367. 10.1093/nar/gki481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeman, D., & Fielding, B. C. (2019). Coronavirus envelope protein: Current knowledge. Virology Journal, 16(1), 1–22. 10.1186/s12985-019-1182-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwede, T. (2013). Protein modeling: What happened to the “protein structure gap”? Structure (London, England : 1993), 21(9), 1531–1540. 10.1016/j.str.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahinozzaman, M., Ishii, T., Ahmed, S., Halim, M. A., & Tawata, S. (2020). A computational approach to explore and identify potential herbal inhibitors for the p21-activated kinase 1 (PAK1). Journal of Biomolecular Structure & Dynamics, 38(12), 3514–3513. 10.1080/07391102.2019.1659855 [DOI] [PubMed] [Google Scholar]
- Shin, W. H., Kang, X., Zhang, J., & Kihara, D. (2017). Prediction of local quality of protein structure models considering spatial neighbors in graphical models. Scientific Reports, 7(1), 40629–40614. 10.1038/srep40629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studer, G., Biasini, M., & Schwede, T. (2014). Assessing the local structural quality of transmembrane protein models using statistical potentials (QMEANBrane). Bioinformatics (Oxford, England), 30(17), i505–511. 10.1093/bioinformatics/btu457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu, K. L., Chan, C. P., Kok, K. H., Chiu-Yat Woo, P., & Jin, D. Y. (2014). Suppression of innate antiviral response by severe acute respiratory syndrome coronavirus M protein is mediated through the first transmembrane domain. Cellular & Molecular Immunology, 11(2), 141–149. 10.1038/cmi.2013.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu, Y. L., Teoh, K. T., Lo, J., Chan, C. M., Kien, F., Escriou, N., Tsao, S. W., Nicholls, J. M., Altmeyer, R., Peiris, J. S. M., Bruzzone, R., & Nal, B. (2008). The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. Journal of Virology, 82(22), 11318–11330. 10.1128/JVI.01052-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, W., Arakaki, A. K., & Skolnick, J. (2004). EFICAz: A comprehensive approach for accurate genome-scale enzyme function inference. Nucleic Acids Research, 32(21), 6226–6239. 10.1093/nar/gkh956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, W., Chen, C., Lei, X., Zhao, J., & Liang, J. (2018). CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Research, 46(W1), W363–W367. 10.1093/nar/gky473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss, D., Pfefferle, S., Drosten, C., Stevermann, L., Traggiai, E., Lanzavecchia, A., & Becker, S. (2009). Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virology Journal, 6(M), 79–13. 10.1186/1743-422X-6-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, B., & Elofsson, A. (2003). Can correct protein models be identified? Protein Science: A Publication of the Protein Society, 12(5), 1073–1086. 10.1110/ps.0236803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., Heer, F. T., de Beer, T. A. P., Rempfer, C., Bordoli, L., Lepore, R., & Schwede, T. (2018). SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Research, 46(W1), W296–W303. 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham, H. (2009). Ggplot2: Elegant graphics for data analysis. Springer-Verlag. 10.1007/978-0-387-98141-3 [DOI] [Google Scholar]
- Wiederstein, M., & Sippl, M. J. (2007). ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research, 35(Web Server), W407–410. 10.1093/nar/gkm290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold, S., Esbensen, K., & Geladi, P. (1987). Principal component analysis. Chemometrics and Intelligent Laboratory Systems, 2(1–3), 37–52. 10.1016/0169-7439(87)80084-9 [DOI] [Google Scholar]
- Yang, J., Anishchenko, I., Park, H., Peng, Z., Ovchinnikov, S., & Baker, D. (2020). Improved protein structure prediction using predicted interresidue orientations. Proceedings of the National Academy of Sciences of the United States of America, 117(3), 1496–1503. 10.1073/pnas.1914677117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Yan, R., Roy, A., Xu, D., Poisson, J., & Zhang, Y. (2015). The I-TASSER suite: Protein structure and function prediction. Nature Methods, 12(1), 7–8. 10.1038/nmeth.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., & Skolnick, J. (2005). TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Research, 33(7), 2302–2309. 10.1093/nar/gki524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobayer, N., & , Hossain, A. A. (2018). In silico Characterization and Homology Modeling of Histamine Receptors. Journal of Biological Sciences, 18(4), 178–191. https://doi.org/ 10.3923/jbs.2018.178.191 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.