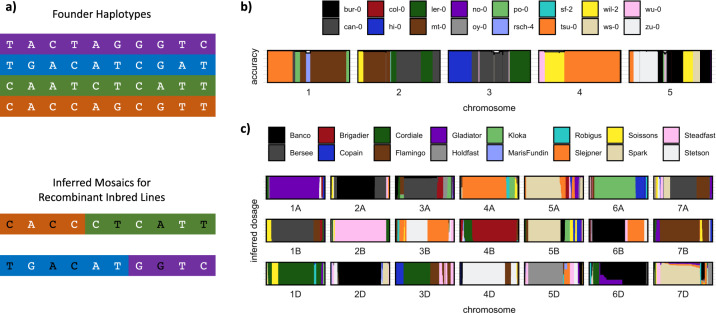
Fig. 3. Reconstruction of MAGIC recombination mosaics and imputation.
a In this schematic example, the MAGIC founders are genotyped more densely and confidently than the MAGIC recombinant inbred lines. The observed genotypes in the recombinant inbred lines (white) can be used to infer the ancestry mosaics (background colours) from which unobserved genotypes (black) can be imputed. Two examples of ancestry mosaics reconstructed from low-coverage sequence data in b Arabidopsis thaliana and c bread wheat. In b, the ancestry mosaic is estimated using the Reconstruction program (http://mtweb.cs.ucl.ac.uk/mus/www/19genomes/MAGICseq.htm), and accuracy is assessed as the fraction of mismatches in each block between the inferred founder haplotypes and calls derived directly from low-coverage sequencing data. In c, the inferred ancestry proportion probabilities (dosages) are emitted after imputation using the software, STITCH (Davies et al. 2016).