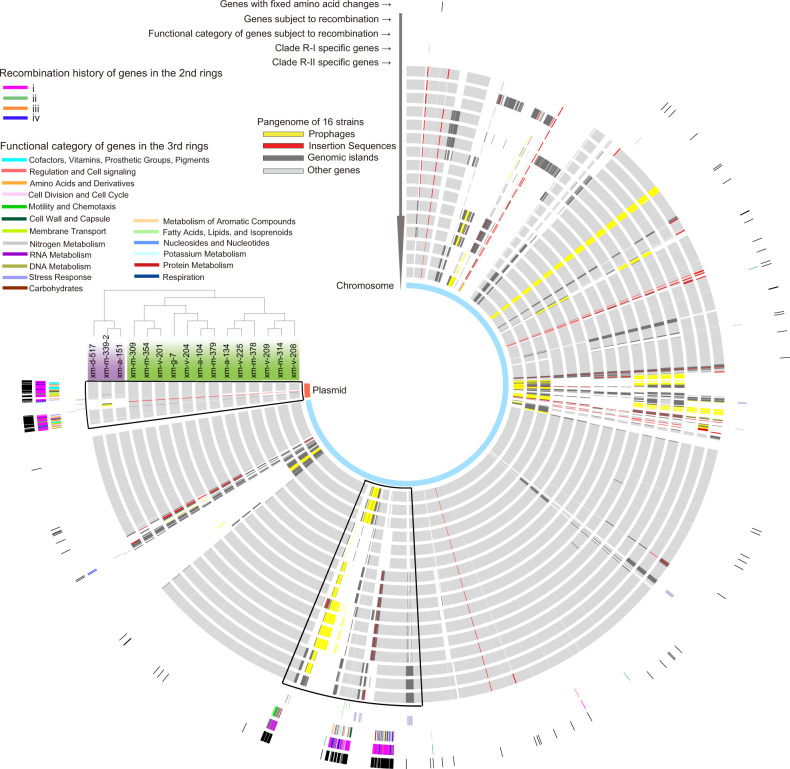
Fig. 2. The pangenome of the Roseobacter population.
From outer to inner rings: (1) core genes with fixed amino acid changes between Clade R-I (taxa shaded in purple) and Clade R-II (taxa shaded in green); (2) core genes (n = 200) subjected to recombination with external lineages. These families are classified into four groups each with a distinct color. Each group represents a distinct history of recombination events illustrated in Fig. S9; (3) SEED subsystem functional category assignment for each of the 200 core genes, with each category shown in a distinct color. Unassigned genes are not represented; (4–5) genes universally and exclusively found in Clade R-I (purple) and Clade R-II (green), respectively; (6–21) genomes of the 16 Roseobacter strains, with the order following the display of these strains in their phylogeny attached to the circos plot. The genome of xm-d-517 is closed, which is used as a skeleton to build the pangenome plot of these 16 strains. The core genes subject to recombination with external lineages are clustered in two adjacent chromosomal regions (framed in one box) and one plasmid region (framed in another box). Mobile genetic elements including prophages (yellow), insertion sequences (red) and genomic islands (dark gray) are mapped to each genome; (22) Chromosome and plasmid are indicated in light blue and light red, respectively.