Fig. 4. WT-based and NifA*-based cocultures exhibit distinct metabolic phenotypes.
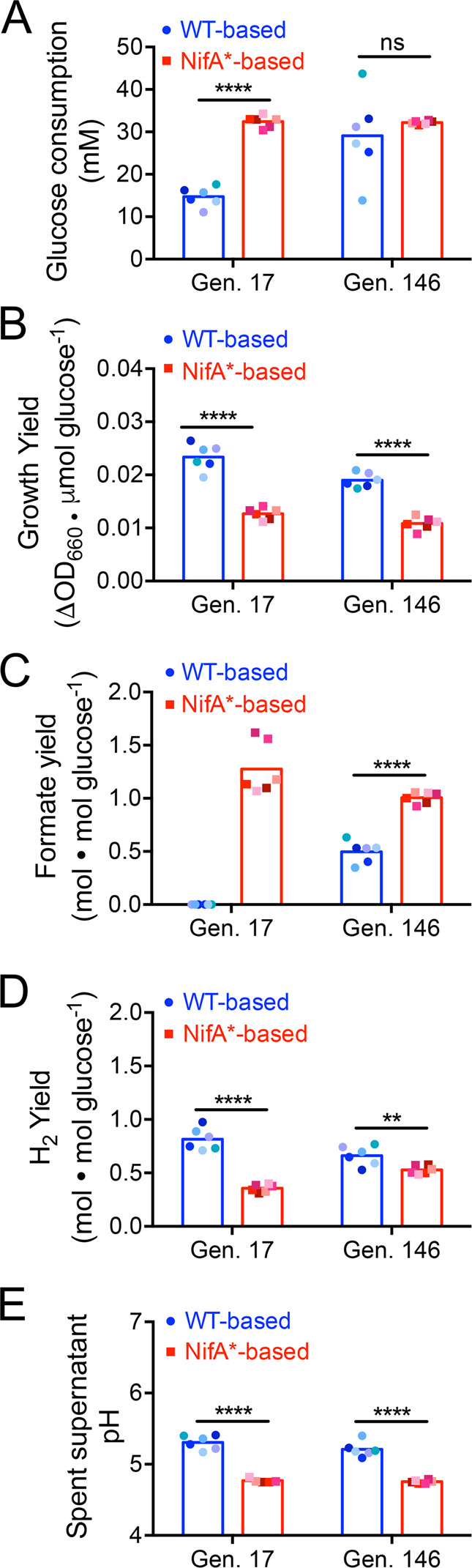
Glucose consumption (a), growth yield (b), formate yield (c), H2 yield (d), and final pH (e) for WT-based (CGA4001, derivative of WT/CGA009 with inactive hydrogenase) and NifA*-based (CGA4003, derivative of NifA*/CGA676 with inactive hydrogenase) revived coculture lineages at generation (Gen.) 17 and 146. Points are single measurements for individual lineages and bars are means, n = 5–6; multiple t-tests with Holm–Sidak post hoc test, **p < 0.01, ****p < 0.0001. Different shades indicate different lineages. Metabolite measurements below detection limit were approximated to be 0. The pH of lineage P at Gen. 17 was not quantified because the culture tube broke prior to measurement.
