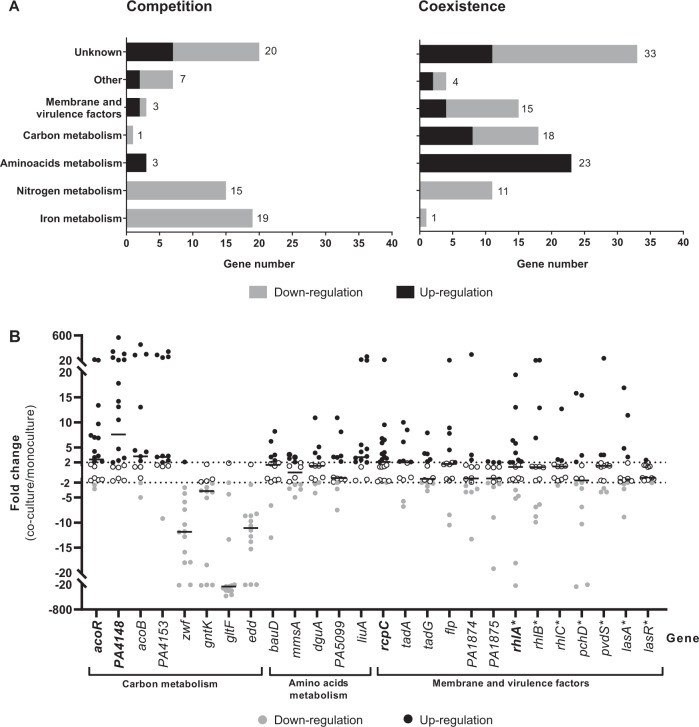
Fig. 1. Alteration of the P. aeruginosa transcriptome induced by co-culture with S. aureus.
a Number of under-expressed (grey bars) and overexpressed (black bars) genes of P. aeruginosa during co-culture with S. aureus for competitive (left) or coexisting (right) pairs. PA2596 competition and PA2600 coexistence strains were cultivated in the absence or presence of SA2597 or SA2599, as described in Supplementary Fig. S1. RNAs were extracted after 4 h of culture and the RNAseq analysis was performed as described in the “Material and methods” section. A gene was considered as differentially expressed when the fold change was >|2log2| with an adjusted P value <0.05. Functional classification was performed using the KEGG database and the literature. b Fold change of 26 P. aeruginosa gene expression induced by co-culture with S. aureus during coexistence interaction. Twenty-one coexisting S. aureus–P. aeruginosa pairs were isolated from separate CF sputa recovered from 20 different patients. Each P. aeruginosa strain was cultivated in the absence or presence of its co-isolated S. aureus strain. RNAs were extracted after 4 h of culture and gene expression was assayed by RT-qPCR. A gene was considered as differentially expressed when the fold change was >|2|, indicated by dotted lines. Black lines indicate the median. Genes were tested in 14 (regular character) or 21 (bold character) strains. The list of the strains used is shown in Supplementary Table S1. Genes annotated with (*) were not identified as dysregulated during the RNAseq experiment.