Abstract
While chromosome 1p36 deletion syndrome is one of the most common terminal subtelomeric microdeletion syndrome, 1p36 microduplications are rare events. Polymicrogyria (PMG) is a brain malformation phenotype frequently present in patients with 1p36 monosomy. The gene whose haploinsufficiency could cause this phenotype remains to be identified. We used high-resolution arrayCGH in patients with various forms of PMG in order to identify chromosomal variants associated to the malformation and characterized the genes included in these regions in vitro and in vivo. We identified the smallest case of 1p36 duplication reported to date in a patient presenting intellectual disability, microcephaly, epilepsy, and perisylvian polymicrogyria. The duplicated segment is intrachromosomal, duplicated in mirror and contains two genes: enolase 1 (ENO1) and RERE, both disrupted by the rearrangement. Gene expression analysis performed using the patient cells revealed a reduced expression, mimicking haploinsufficiency. We performed in situ hybridization to describe the developmental expression profile of the two genes in mouse development. In addition, we used in utero electroporation of shRNAs to show that Eno1 inactivation in the rat causes a brain development defect. These experiments allowed us to define the ENO1 gene as the most likely candidate to contribute to the brain malformation phenotype of the studied patient and consequently a candidate to contribute to the malformations of the cerebral cortex observed in patients with 1p36 monosomy.
Subject terms: Gene regulation, Genetics research
Introduction
Rearrangements of the terminal region of chromosome 1 are particularly frequent and represent up to 20% of all unbalanced rearrangements reported to cause intellectual deficiency and birth defects in the human genome [1]. Terminal deletions of chromosome 1 account for up to 67% of all rearrangements on this chromosome [2] and are causing one of the most frequent subtelomeric microdeletion syndrome: 1p36 monosomy (MIM 607872) with a frequency of 1 in 5000 [3]. No common breakpoints have been identified and these terminal deletions have a highly variable size, ranging from 1 to 12.9 Mb [4, 5]. The severity of the phenotype is generally correlated to the size of the deletion and most patients with 1p36 monosomy suffer from intellectual disability, growth, cardiac, and hearing defects, facial dysmorphism, hypotonia, epilepsy, and microcephaly [6, 7].
The widespread usage of high-resolution array-based comparative genomic hybridization (CGH) in clinical settings has revealed smaller interstitial deletions in 1p36 [8–10]. The molecular characterization of these small rearrangements has allowed to correlate the clinical signs of the patients with the gene content of the deleted segments. Using this strategy, several candidate genes were proposed such as KCNAB2 for epilepsy [11], SKI for cleft palate [12], MMP23 for abnormal cranial suture closure [13] or GABRD for the neuropsychiatric and neurodevelopmental phenotypes [14]. However, there are discrepancies on some phenotype genotype correlations [9] and a possibility exists that position effects could also cause part of the 1p36 phenotype whereby genes located outside of the deleted segments will cause some of the observed phenotypic characteristics [8].
Although nonallelic homologous recombination was proposed as a mechanism for the 1p36 deletions [2, 15], there are very few reported duplications of this region. Only two cases of isolated duplications have been reported [16, 17] in addition to duplications associated with more complex rearrangements [18, 19]. Duplications are more difficult to identify using classical cytogenetic techniques and 1p36 duplications might cause a milder phenotype [16]. Given the small number of duplications identified in 1p36 and their heterogeneity, it is currently not possible to define a syndrome and the only constant phenotypic traits in carriers are nonspecific features such as microcephaly, developmental delay, and growth retardation [19].
Here, we report one of the smallest 1p36 duplication identified to date. This rearrangement occurred in a patient presenting with intellectual deficiency, microcephaly, epilepsy, and bilateral perisylvian polymicrogyria (PMG). The same patient also presents a small 12p13.1 duplication. PMG has frequently been described in patients with the 1p36 deletion syndrome [20, 21] but the gene responsible for this phenotype has not been identified. Conversely, no PMG has ever been described in patients carrying a rearrangement of the 12p13.1 region [20]. We thus focused our analysis on the 1p36 region. We characterized the rearrangement in detail, performed in situ hybridization to describe the developmental profile of expression of the two genes contained in the duplicated region in 1p36 and we studied the consequences of the inactivation of these two genes in vivo using in utero electroporation. These experiments allowed us to define the enolase 1 (ENO1) gene as the most likely contributor to the brain malformation phenotype of the studied patient and a candidate contributor to the malformations of the cerebral cortex observed in patients with 1p36 monosomy.
Materials and methods
Our project obtained ethical approval from Comité de Protection des Personnes Sud-Méditerranée and signed informed consent was obtained from study participants.
Array CGH
Array CGH was performed using NimbleGen 3 × 720K Whole Genome arrays (Roche Nimblegen, Madison, WI), providing an average resolution of 2.5 kb. The microarrays were hybridized and scanned on an Agilent scanner. Data were then extracted using NimbleScan software v2.5 and analysed with SignalMap v1.9 software. Regions showing an abnormal dosage were compared with regions already known to represent nonpathogenic copy number variations (CNV) listed in the database of genomic variants (http://dgv.tcag.ca/gb2/gbrowse/dgv2_hg38). CNVs that were totally included in already reported regions in at least two unrelated healthy individuals, using non-BAC arrays, were considered probably benign and were not studied further. The genomic variant was submitted to the Global Variome shared LOVD and is accessible at the following URL: https://databases.lovd.nl/shared/individuals/00285820.
Classical and quantitative PCR
Classical PCR was used to determine the orientation of the duplicated segment using primers designed at each extremity of the duplicated segments, in both orientations. Expression analysis was performed using cDNAs prepared with RNAs obtained from different human tissues (BD Biosciences, Palo Alto, California, USA) or total RNA of patient and healthy control individuals extracted from lymphoblasts using TRIzol reagent (Invitrogen Corp., Carlsbad, CA, USA). Reverse transcription (RT) reactions were performed using 5 μg total RNA. PCR reactions were performed in 20 μl of Superscript II reaction buffer (Invitrogen, Carlsbad, California, USA) containing 0.01 M dithiothreitol, 7.5 ng/μl of dN6, 20U of RNase inhibitor (Invitrogen), 10 mM dNTP and 200 U of Superscript II reverse transcriptase (Invitrogen) for 1 h at 42 °C. All classical PCR reactions were performed using the T1 thermocycler (Whatman-Biometra, Goettingen, Germany) in a total volume of 50 μl, containing 1X PCR buffer, 0.2 mM dNTPs, 1.5 mM MgCl2, and 1U Taq polymerase. Real-time PCR reactions on genomic DNA or cDNA were performed using the LightCycler 480 system (Roche, Indianapolis, IN, USA) using the SYBR Green I Master Kit (Eurogentec, Seraing, Belgium) with 2 μl of cDNA and 200 nM of each PCR primer. Each reaction was performed in triplicate. All primer sequences are detailed in Supplementary Table 1.
Fluorescent in situ hybridization
For FISH analysis, BAC (RP11–651L10 on 1p36.23) and fosmid (G248P81726 on 12p13.1). DNAs were labeled with Spectrum Orange d-UTP using a nick translation kit (Abbott Diagnostic, Rungis, France) according to the instructions of the manufacturers. Hybridization was performed using the standard procedures [22]. Chromosomes were counterstained with 4,6-diamino-2-phenylindole (DAPI) and observed using an Axioplan-2 Zeiss fluorescent microscope (Zeiss) and the images captured with a CCD camera (Photometrics « SenSys »). Information was collected and merged using IPLab Spectrum software (Vysis).
In situ hybridization
In situ hybridization was performed on embryonic and adult mouse tissues using specific antisense riboprobes. Embryos were fixed by immersion in 0.1 M phosphate-buffered saline (PBS), pH7.4, containing 4% paraformaldehyde (2–8 h at 4 °C). Adult mice used for in situ hybridization were transcardially perfused (chilled saline for 1 min followed by 0.1 M PBS containing 4% paraformaldehyde for 10 min), then embryos or brains were collected and fixed by immersion in 0.1 M PBS, pH7.4, containing 4% paraformaldehyde (24 h at 4 °C). Tissues were cryoprotected overnight in PBS containing 20% sucrose and embedded in Optimal Cutting Temperature compound (VWR, Fontenay-sous-Bois, France) before freezing. The blocks were then stored at −80 °C until use. Cryostat sections (12 mm) were collected on Superfrost+ slides and stored at −80 °C until used. The antisense and sense riboprobes used in this study correspond to nucleotides 149,622,695–149,622,919 for Eno1 and 149,874,114–149,944,365 for Rere on mouse chromosome 4, and to nucleotides 135,023,416–135,023,736 for Ddx47 on mouse chromosome 6 (NCBI m37). In situ hybridization was performed as previously described [23] with antisense probes transcribed with T7 or SP6 polymerases, and the digoxigenin-labeled probe was detected using an antidigoxigenin antibody coupled to alkaline phosphatase in the presence of NBT/BCIP (Roche, Basel, Switzerland). Control experiments were performed using corresponding sense riboprobes on adjacent sections, giving either no signal or a uniformly low background as expected.
In utero electroporation of small hairpin RNAs (shRNAs)
shRNA sequences were designed using sequence of the common regions of the 3′UTR and 5′UTR of rat and mouse transcripts. They correspond to positions 1659_1680 and 1668_1688 of transcript ENSRNOT00000024106 for shEno1–1 and shEno1-2, respectively and positions 852_874 and 6227_6249 of transcript ENSRNOT00000024443 for shRere-1 (5′UTR) and shRere-2 (3′UTR), respectively. The fragments were amplified using PCR and cloned into the mU6pro vector [24]. Inserts were sequenced to confirm the absence of the variant. In order to test the efficacy of the shRNAs, rat 3T3 cells were transfected with each shRNA and the levels of Eno1 or Rere expression were measured using quantitative PCR, taking into consideration the percentage of transfected cells.
Electroporations were performed at E15.5 in WISTAR rats (Janvier, Le Genet Saint Isle, France). Pregnant females were anesthetized with a mixture of ketamine (100 mg/kg) and Xylazine (10 mg/kg) and the uterine horns were exposed. 0.5–1 μl of DNA solution was injected into one lateral ventricle of each embryo using pulled glass capillaries and a microinjecter (Picospritzer ΙΙ). The DNA solution contained 0.5 mg/ml of a pCAG-RFP plasmid either alone or with 1.5 mg/ml of shRNA, with fast green 2 mg/ml (Sigma). Embryo heads across the uterine wall were placed between tweezer-type electrodes (NEPA Gene). Five electric pulses (50 V; 50 ms) were delivered at 950 ms intervals, using an electroporator BTX ECM 830 (BTX Harvard Apparatus). Uterine horns were placed back and the abdomen sutured. Rat embryos were deeply anaesthetized with pentobarbital (100 mg/kg) and transcardially perfused (with 4 % paraformaldehyde for 10 min). The brains were removed, post-fixed for 5 h, and cut into coronal sections using a vibratome (Leica). Sections were permeabilized (0.1 % Triton X-100 PBS), and labeled with 4,6-Diamino-2- phenolindole dihydrochloride (DAPI), in order to visualize the nuclei.
Cells dissociation and sorting
Brains were extracted 3 days after coelectroporation with either shEno1, shRere, or shScramble and pCAG-GFP plasmids. Subventricular zones and cortical plates were dissected manually. Cells were dissociated using the MACs « Adult Brain Dissociation Kit, mouse, and rat » (Miltenyi Biotec, Paris, France). Cells were gated based on forward scatter, side scatter, and trigger pulse width. Autofluorescent cells were excluded based on their nonspecific signal in the Amcyan channel. Cells were further gated as GFP+ (Green Fluorescent Protein). Cells were sorted using FACS (Fluorescence-Activated Cell Sorting) into lysis buffer (10:1 mix of Resuspension Buffer and Lysis Enhancer from Cells Direct one-step qRT-PCR kit, Thermofisher, Massachusetts, USA). After sorting, tubes were immediately frozen on dry ice, then stored at −80 °C until processing for RT-qPCR. RT-qPCR results were the means of 3 wells per condition per gene.
Results
Case report
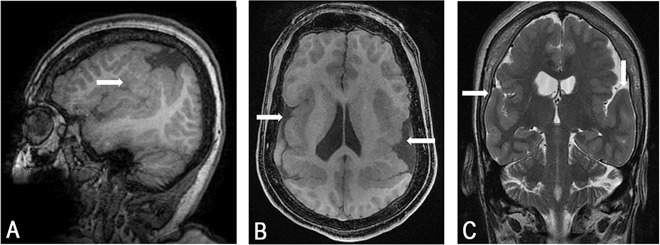
The patient, a 35 years old woman, was born at 32 weeks after uneventful delivery. From 8 months of age developmental delay was noticed gradually becoming consistent with bilateral opercular syndrome including severe motor dysphasia, difficulties in chewing and swallowing, continuous drooling, accompanied by hemiparesis, microcephaly, and progressive kyphoscoliosis. Although cognition was not formally assessed, the patient showed severe expressive speech dyspraxia and was studying in a special school system. She was never independant in her daily skills and had poor communication skills. She could write simple words, could communicate with communication board using more than 100 signs and was able to do calculation up to 10. Seizures started at 3 years of age and included GTCs, absences, and drop attacks. Repeated EEG’s showed bilateral synchronous and independent slow spike wave discharges leading to the diagnosis of symptomatic Lenox-Gastaut syndrome. Seizures were and are resistant to all available anti-epileptic drugs, ketogenic diet but show partial response to vagal nerve stimulation. Currently the woman is wheelchair bound because of severe kyphoscoliosis, recurrent drop attacks, and progressive spasticity. Recurrent aspirations related to drooling and abnormal chewing and swallowing led to G-tube insertion. No cognitive deterioration is noted. MRI showed abnormal temporo-parietal gyration which is consistent with bilateral polymicrogyria (Fig. 1).
Fig. 1. Magnetic resonance imaging of the patient brain showing bilateral perisylvian polymicrogyria (arrows).
Representative sagittal fast spoiled gradient echo (FSPGR) image (a), axial FSPGR image (b) and coronal fast spin echo (FSE) T2 image (c) sections are shown. Notice asymmetry of location and extent of abnormal cortex on both hemispheres and asymmetric ventricles possibly explaining the patients’ hemiparesis in addition to her opercular symptoms and signs.
Identification and characterization of the 1p36.23 and 12p13.1 duplications
We have studied the patients’ DNA using high resolution CGH on arrays providing a mean resolution of 2.5 kb. This analysis detected 33 common copy number variants (CNV) and 5 CNVs that had not previously been reported (Supplementary Table 2). One CNV (loss 1q21.2) was located in an intergenic regions. We focused on candidate protein-coding genes directly impacted by the rearrangements.
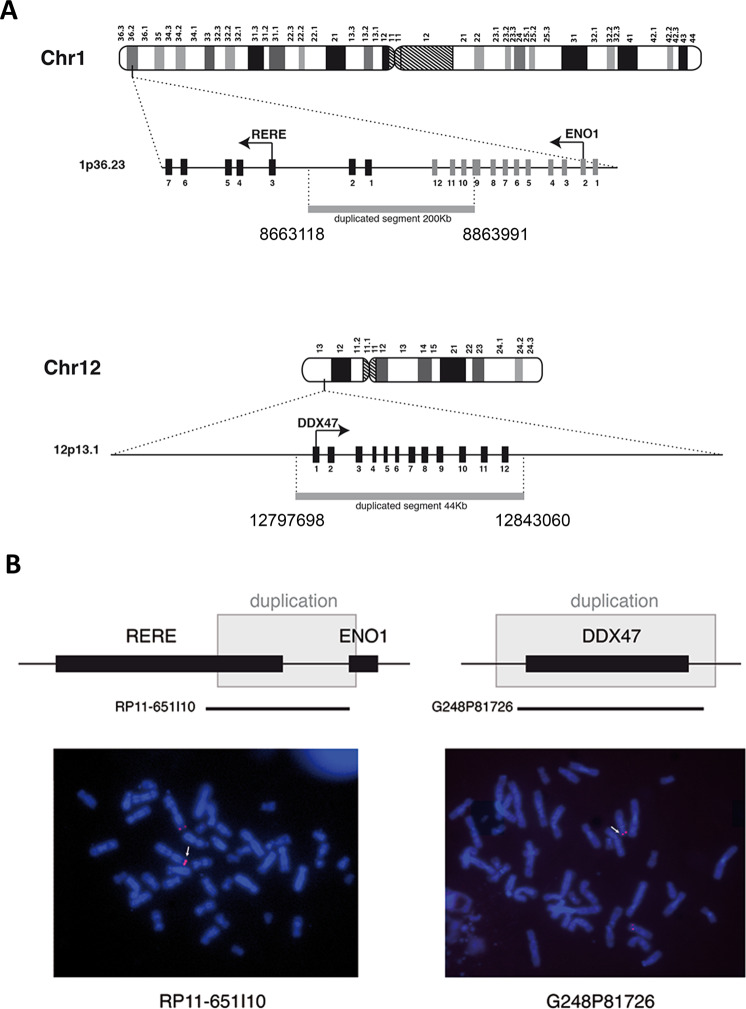
The four remaining CNV (Supplementary Table 2) contain transcription units. Using quantitative PCR on genomic DNA of the patient and her mother (the paternal DNA is not available), we showed maternal inheritance for two of them (Supplementary Table 2) and they were not considered further. The last two correspond to duplications that were not inherited from the mother: a 200 kb duplication in 1p36.23 and a 44 kb duplication in 12p13.1, arr[GRCh38]1p36.23(8654490_8663239)_(8864067_8868341)x3, 12p13.1(12789530_12797985)_(12842155_12855828)x3 (Fig. 2a).
Fig. 2. Genomic location and size of the duplications.
a Schematic representation of the genomic duplications identified in the patient on chromosomes 1 and 12, following human genome build 38 (GRCh38). Gray bars show the duplicated regions, based on the first and the last duplicated oligonucleotides on the CGH array. Exons are represented as black or gray boxes and an arrow indicates the position of the translation initiation codon and the orientation of transcription. b Fluorescent in situ hybridization performed on metaphases of the patient with probes RP11-651L10 (BAC clone on chromosome 1) and G248P81726 (fosmid clone on chromosome 12), both spanning the duplicated regions. These probes appear as red signals and white arrows show the duplicated chromosomes.
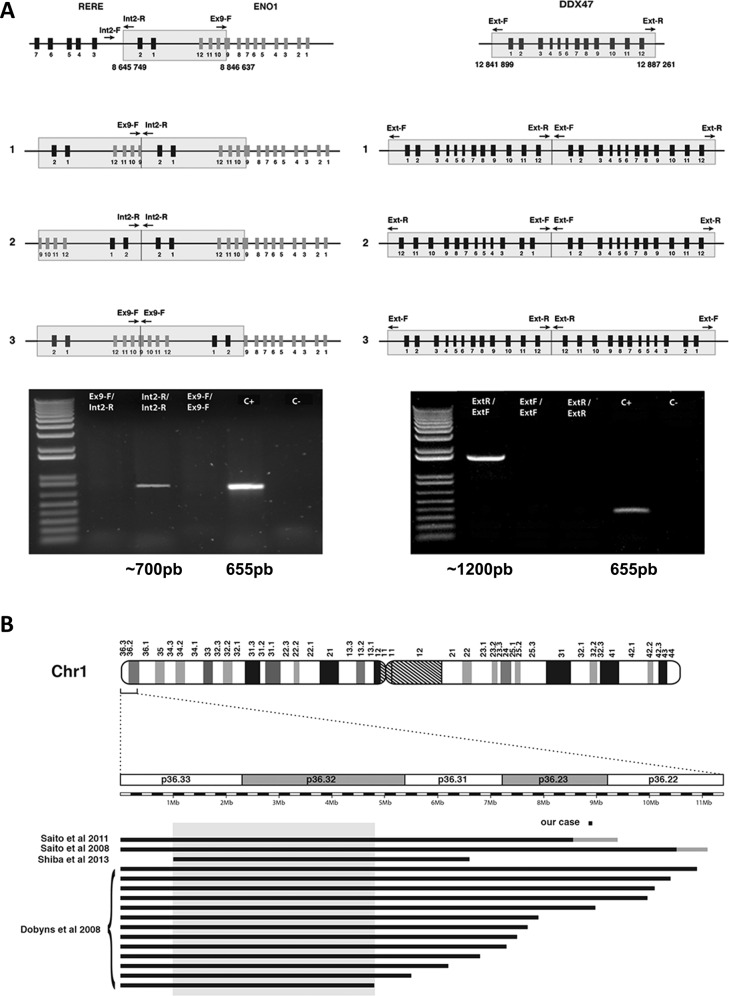
In order to identify the chromosomal location of the duplicated fragments in the patient cells, we performed fluorescent in situ hybridization (FISH) using bacterial artificial chromosomes (BAC) and fosmid probes. This analysis revealed that the two duplication events were intrachromosomal and were located at the original locus (Fig. 2b). Next, we designed a series of PCR primers in order to amplify the junction fragments of the two duplication events and to determine the orientation of the duplicated segments (Supplementary Table 1). We amplified also a part of the TBC1D24 gene like a positive control for the quality of the DNA. This PCRs gave us a band around 700 pb for the 1p36 duplication and a band around 1200 pb for the 12p13.1 duplication. These bands were sequenced and this strategy allowed us to precisely map the duplications breakpoints (Fig. 3a) and to show that the 1p36.23 region was duplicated in mirror while the 12p13.1 region was duplicated in tandem. The 1p36.23 duplication starts at nucleotide 8,663,118 and ends at nucleotide 8,863,991 of chromosome 1 (GRCh38 Hg38). A comparison with previously published cases reveals that this region is the smallest 1p36 rearrangement reported to date (Fig. 3b). The 12p13.1 duplication starts at nucleotide 12,797,698 and ends at nucleotide 12,843,060 on chromosome 12 (GRCh38 Hg38).
Fig. 3. Structure of the duplicated segments and comparison with published cases.
a Determination of the orientation of the duplicated segments. The three possible configurations are shown: in tandem (1), in mirror at the distal side (2) or in mirror at the proximal side (3). Arrows indicate orientation of PCR primers. Positive PCR amplifications reveal that the 1p36 duplication occurred in mirror while it occurred in tandem in 12p13.1. The positive control corresponds to the amplification of a portion of the TBC1D24 gene on chromosome 16, to check for proper PCR conditions. b Location of the 1p36 duplicated region in the reported patient with respect to previously reported and precisely characterized 1p36 deletion cases with PMG. Deleted regions are represented as black lines, gray lines identify uncertainly delimited deletions. The gray box denotes the previously published minimal critical regions for PGM, located between 1 and 4.8 Mb on chromosome 1.
We next considered the gene content of the two duplicated regions. The duplicated segments contain the RERE and ENO1 genes on chromosome 1 and the DDX47 gene on chromosome 12. Based on the characterization of the duplication breakpoints performed above, we observed that the RERE and ENO1 genes had been interrupted by the chromosome 1 duplication while the DDX47 gene was entirely contained within the duplication on chromosome 12. We used a panel of mRNAs prepared from various human tissues to detect ENO1, RERE, or DDX47 transcripts. Because several alternative transcripts exist for the three genes, we designed PCR primers allowing the amplification of all isoforms (Supplementary Table 1). We found that the three genes have a ubiquitous expression (Supplementary Figure S1A). Given that the three transcripts were detected in lymphocytes, we used quantitative PCR to quantify the expression levels of the three transcripts using mRNA prepared from lymphoblasts of the patient (Supplementary Table 1). This analysis revealed that the ENO1 gene was under-expressed (54% of wild type level) in the cells of the patient while DDX47 was overexpressed (216% of wild type level) (Figure S1B). We were not able to quantify the expression level of RERE because we observed that the expression of this gene is highly variable in control cell lines (data not shown).
Expression of ENO1 and RERE in mouse development
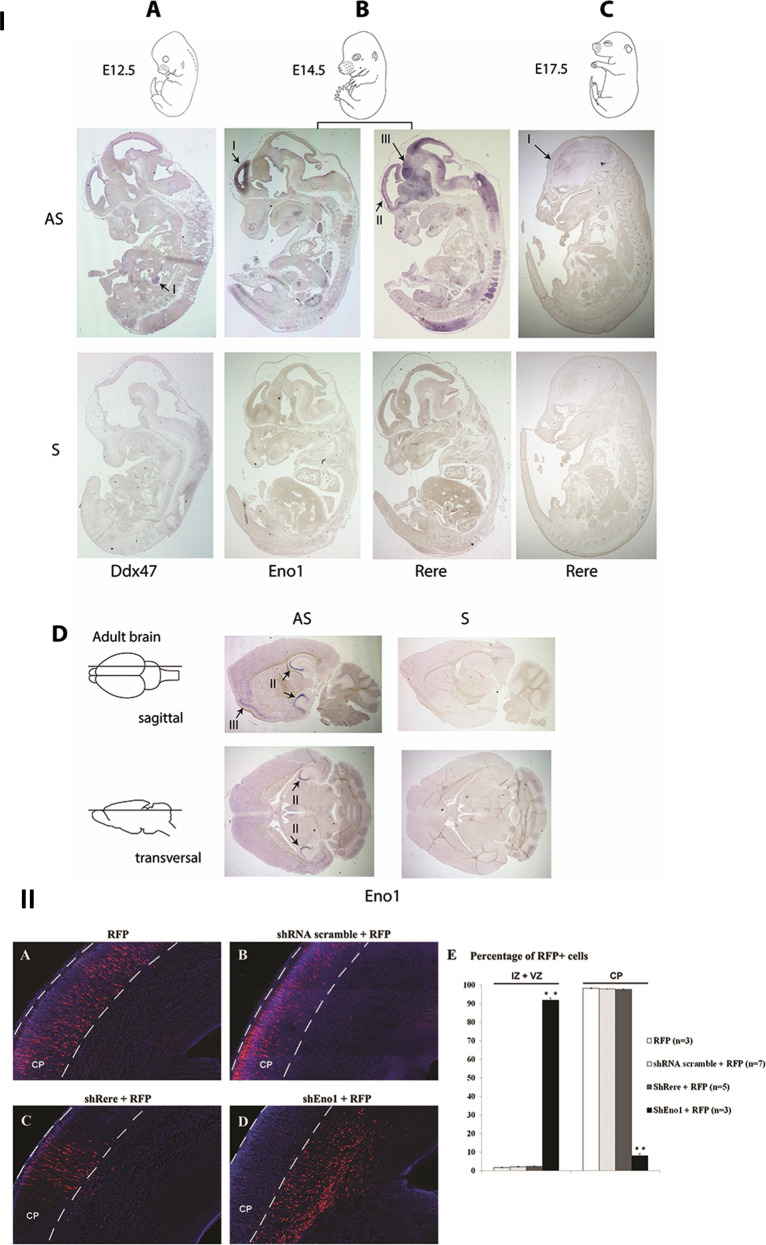
In order to document the spatio-temporal expression of the three genes, we used in situ hybridization in mouse embryos and in the adult mouse brain (Fig. 4I). During the development, Eno1 is expressed at embryonic stage E14.5 in the inner (ventricular) side of the neopallial cortex (Fig. 4I.b). The expression of this gene subsequently decreases after E14.5. In the adult brain, we detected a strong expression of Eno1 in the cornus ammoni (CA) regions CA2 and CA3 of the hippocampus and a moderate expression in the cerebral cortex (Fig. 4I.d). The Rere mRNA was detected at E14.5 in the central nervous system, with a strong expression in the neopallial cortex and the midbrain (Fig. 4I.b). The expression in the neopallial cortex appears to be higher in the outer (pial) side than the inner (ventricular) side. The transcripts of Rere are detected to a lesser extent at later embryonic stages (E17.5) (Fig. 4I.c).
Fig. 4. In situ hybridization and in utero electroporation.
I. Expression of Eno1, Rere, and Ddx47 in mouse development using in situ hybridization. a–c Sagittal sections of mouse embryo at E12.5 (a), E14.5 (b), and E17.5 (c). d Sagittal and transverse sections of adult brains. Eno1 was strongly expressed in the inner ventricular side of the neopallial cortex at E14.5 (b–I), in the CA2 and CA3 fields of hippocampus and in the cerebral cortex in the adult brain (d-II and d-III, respectively). Rere expression was detected in central nervous system and strongly in the neopallial cortex (outer side) (b-II) and the midbrain (b-III) at E14.5. At E17.5, Rere expression was also detected in the neopallial cortex but at a lower level (c-I). A faint expression of Ddx47 transcript was detected in the urogenital region at E12.5 (a-I). II. Coronal sections of rat brain at E20, five days after electroporation. The presence of RFP alone (a), of a scramble shRNA (b), or shRere-2 (c) does not disturb neuronal migration. The presence of shEno1-1 severely impairs neuronal migration (d). Cells were counted in the different regions of the developing brain: intermediate (IZ) or ventricular (VZ) zones, or the cortical plate (CP) and plotted for comparison (E). Results are reported as mean ± standard error of the mean (SEM), and Student t test was used to test statistical significance, p value = 0.0008 for the significative difference in IZ and VZ and p value = 0.0009 for the significative difference in CP. A p value < 0.001 was considered to be statistically significant. **p < 0.001.
The Ddx47 transcripts were difficult to detect in the different development stages. We observed a faint mRNA expression in the urogenital region at E12.5 (Fig. 4I.a). No signal was observed in the central nervous system, neither at any embryonic stages nor in the adult CNS.
Neuronal migration evaluation
In order to observe the consequences for brain development of a downregulation the two genes located in 1p36, we used in utero electroporations in rat embryos. We first designed several small hairpin (sh) RNAs targeting the two genes (shEno1-1 and shEno1-2, shRere-1, and shRere-2). These shRNAs were designed to target the 3′ and/or 5′ untranslated region (UTR) of the rat Eno1 and Rere transcripts because subsequent experiments were performed in that species. In order to test the efficacy of the shRNAs, rat 3T3 cells were transfected with each shRNA and the levels of Eno1 or Rere expression downregulation were measured using quantitative PCR, taking into consideration the percentage of transfected cells. We selected the most efficient shRNAs for each gene: 69.2 and 45% reduction of Eno1 expression compared with wild type levels for shEno1-1 and shEno1-2 respectively, 4,8% and 49,7% reduction of Rere expression compared with wild type levels for shRere-1 and shRere-2, respectively.
Next, we performed in utero electroporations with either the shEno1-1 shRNA or shRere-2 and a red fluorescent protein (RFP) plasmid or the RFP plasmid alone. Embryos were electroporated at E15.5. Embryos were analysed at E20, 5 days post-electroporation. In embryos electroporated with the RFP plasmid alone, with the scramble shRNA or with shRere, 98.3% (SEM = 0.24%; n = 3 brains), 97.9% (SEM = 0.1%; n = 7 brains) and 97.7% (SEM = 0.31%; n = 5 brains) of electroporated neurons reached the cortical plate, respectively (Fig. 4II.a, 4II.b, 4II.c). On the contrary, in embryos electroporated simultaneously with shEno1-1 and the RFP plasmid, only 8% of electroporated neurons (SEM = 1.18%; n = 3 brains) reached the cortical plate and a large number of RFP positive neurons were still visible in the intermediate zone (IZ) at E20 (Fig. 4II.d). A summary of the results of these experiments is presented in Fig. 4II.e.
In order to test the efficacy of the shEno1-1 and the shRere-2 in vivo, we performed in utero electroporation in Wistar rat embryos using either the shEno1-1 or shRere-2 and a Green Fluorescent Protein (GFP) plasmid (pCAG-GFP) or the GFP plasmid alone. Embryos were electroporated at E15.5. In order to see the effect of each shRNA on gene expression during the cell migration, brains were extract at E18, 3 days post-electroporation. Cells were dissociated from 2 shScramble (control), 10 shEno1, and 11 ShRere dissected brains. After dissociation, we FACS sorted cells based on the GFP+ fluorescence compared with GFP negative control cells. After the FACs sorting, we tested the expression of Eno1 and Rere by RT-qPCR (Supplementary Table 1). The expression of the Eno1 gene was reduced by 41% after the electroporation of the shEno1 however the expression of the Rere gene was reduced by 56% after the electroporation of the shRere (Supplementary Figure S2). In conclusion, the downregulation of the Rere gene expression by 56% in the electroporated neuroblasts does not perturb their migration to the cortical plate, however, the downegulation of the Eno1 gene expression by 41% in the electroprated neuroblasts disturbs their migration to the cortical plate and blocks 92% of them in the IZ.
Discussion
The search for chromosomal rearrangements in patients presenting a malformation of the cerebral cortex is a powerful approach to identify new candidate genes. We used this strategy to study a patient presenting bilateral polymicrogyria. PMG is a malformation of the cerebral cortex characterized by a loss of the normal microscopic and macroscopic architecture with the appearance of numerous small and unfolded gyri and disorganization of cortical layering [20]. PMG can be caused by genetic variants. It is genetically heterogeneous and variants in several genes were reported to be the cause of the malformation [25].
We then identified two duplications in our patient. Despite that duplication could change the structure of the genome, the topologically associated domains organisation and therefore the gene expression [26], we focused on protein-coding genes directly impacted by the rearrangements. The first duplication concerns the 12p13.1 chromosomal region and contains a single gene called DDX47, entirely duplicated. This gene encodes a poorly described DEAD box protein member having a putative role as an RNA helicase [27]. We found that DDX47 is overexpressed in the lymphoblasts of the patient, as expected following the duplication of the entire transcription unit. However, expression analysis in mouse embryonic development shows that the DDX47 transcript is mainly detected in the urogenital system. The absence of similar cases in genomic variant databases, the absence of linkage to 12p13 of a disease associated with a malformation of the cerebral cortex, combined to the expression pattern of DDX47, did not supported an obvious role for this transcript in cortical development and we did not studied this gene further. The second duplication concerns the 1p36.23. This small 200 kb duplication contained two interesting genes, ENO1 and RERE. Sequencing revealed the presence of two similar regions surrounding the duplication which could be the cause of duplication following homologous recombination. The first region is located between the base pairs 8663091 and 8663103 (CTATTTGGGCGGG) of the RERE gene and the second region is located between the base pairs 8863979 and 8863991 (CTCTTTGGGTTGG) of the ENO1 gene.
We showed that ENO1 expression was reduced by 50% in the patient cells mimicking the haploinsufficiency caused by a monosomy. Expression analysis in mouse development revealed a strong expression of both transcripts in the central nervous system. Interestingly, 1p36 monosomy has been associated with bilateral polymicrogyria [20, 21]. Although 1p36 deletions are common, early descriptions did not provide cerebral imaging results [5, 28–30]. More recently however, correlations between 1p36 rearrangements and polymicrogyria were reported. In a large study of 64 patients presenting a deletion of 1p36 [20], 20% of the cases had polymicrogyria. Combined with three additional reports [3, 21, 31], these studies allowed to define a putative minimal critical region of 3.8 Mb in 1p36 for polymicrogyria localized between 1 Mb and 4.8 Mb from the 1p telomere (see Fig. 3b). The 1p36 rearrangement described here is the smallest identified to date in association with polymicrogyria. Although it lies outside of the minimal critical region defined previously [20], it contains the RERE gene, proposed to be a candidate gene for polymicrogyria [21]. RERE encodes a member of the arginine/glutamic acid repeat-containing protein family. This protein was shown to be a nuclear receptor coregulator interacting with NR2F2 and NR2E1 [32, 33] and to interact with histone deacetylases in the mouse embryo [31, 34], but its function is not completely elucidated [34]. Different mouse models with null or hypomorphic RERE alleles have been produced [35–37]. These models revealed that a deficiency of RERE leads to a wide range of developmental defects including the brain, heart, and kidney. One of these was described with abnormal cerebellum development [37] but without neuronal migration defects in the cerebral cortex. Moreover, in human, a new genetic syndrome was recently described in RERE variant cases [38, 39]. It is characterized by neurodevelopmental disorder that may be accompanied by brain, eye, and/or heart anomalies, but none of the 19 individuals reported in the Jordan et al. and Fregeau et al. studies had any kind of polymicrogyria.
We were not able to assess the expression level of RERE in the cells of our patient because of the large variability of RERE expression in normal human lymphoblasts. However, taking into account the absence of neuronal migration defect in mouse models or in human RERE syndrome, as well as in our own in utero electroporation experiments, we do not favor a role of RERE variants in the polymicrogyria phenotype observed in the duplicated patient. The absence of a neuroblasts migration defect in heterozygous Rere+/− mice is comparable to our results obtained with the shRere decreasing Rere expression by 50.3%. Nonetheless, RERE deficient embryos that carry both a null or a hypomorphic allele of Rere have been described to have Purkinje cell migration defects [35, 37].
Rather, our results indicate that ENO1 encoding α-enolase, is a good candidate to contribute to that phenotype. Enolase is a glycolytic enzyme involved in production of ATP. The functional enzymes are dimers composed of subunits encoded by separate genes designated ENO1, ENO2, and ENO3 for α, γ, and β subunits, respectively. Type αα-enolase is the predominant form and is found in wide range of tissues. In the developing brain, α dimers switch toward γ-enolase subunits expression while neurons undergo terminal differentiation [40].
The ENO1 transcript levels were reduced by 50% in the cells of the patient with the 1p36 duplication, mimicking monosomy. It is worth noting that 8 heterozygous loss of function variants (excluding the splice variants that can sometimes preserve a reading frame) are found in the 120,292 alleles of the control population in the Genome Agreggation Database (gnomAD). This may suggest that ENO1 dysfunction could contribute to a neuronal migration defect in combination with other factors, rather than acting as a classical monogenic disease causing gene. Should this be true, ENO1 haploinsufficiency would contribute to an oligogenic phenotype. Also, it is possible that the minimal critical region defined in 2008 by Dobyns et al. [20] contains regulatory elements acting on ENO1 and/or other distant transcripts.
The analysis of its expression in mouse development revealed a strong expression in the cerebral cortex during embryogenesis compatible with a role during brain development. Most interestingly, in utero electroporation of interfering Eno1 shRNAs revealed that the migration of electroporated neurons is severely impaired during the early phases of cerebral cortex development. According to the previously mentioned oligogenic hypothesis, an interaction between Eno1 and Rere could participate in polymicrogyria, even though the downregulation of Eno1 alone prevents 92% of neuroblasts from reaching the cortical plate and causes a severe abnormal migration phenotype. Taken together, these elements strongly suggest that ENO1 must be considered as a contributor gene to the phenotype of malformation of the cerebral cortex in the 1p36 region and that it could be screened for variants in patients with polymicrogyria of unknown origin.
Supplementary information
Acknowledgements
We thank the Centre de Ressources Biologiques of La Timone Children’s Hospital (Karine Bertaux, Cécile Mouradian and Annachiara Desandre Giovannoli) for assistance with the samples used in this study and Virginie Bascunana and Dr. Françoise Watrin for technical assistance.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-020-0659-z) contains supplementary material, which is available to authorized users.
References
- 1.D’Angelo CS, Gajecka M, Kim CA, Gentles AJ, Glotzbach CD, Shaffer LG, et al. Further delineation of nonhomologous-based recombination and evidence for subtelomeric segmental duplications in 1p36 rearrangements. Hum Genet. 2009;125:551–63. doi: 10.1007/s00439-009-0650-9. [DOI] [PubMed] [Google Scholar]
- 2.Saito S, Kawamura R, Kosho T, Shimizu T, Aoyama K, Koike K, et al. Bilateral perisylvian polymicrogyria, periventricular nodular heterotopia, and left ventricular noncompaction in a girl with 10.5-11.1 Mb terminal deletion of 1p36. Am J Med Genet A. 2008;146A:2891–7. doi: 10.1002/ajmg.a.32556. [DOI] [PubMed] [Google Scholar]
- 3.Gajecka M, Saitta SC, Gentles AJ, Campbell L, Ciprero K, Geiger E, et al. Recurrent interstitial 1p36 deletions: evidence for germline mosaicism and complex rearrangement breakpoints. Am J Med Genet A. 2010;152A:3074–83. doi: 10.1002/ajmg.a.33733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heilstedt HA, Ballif BC, Howard LA, Lewis RA, Stal S, Kashork CD, et al. Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Am J Hum Genet. 2003;72:1200–12. doi: 10.1086/375179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greco M, Ferrara P, Farello G, Striano P, Verrotti A. Electroclinical features of epilepsy associated with 1p36 deletion syndrome: a review. Epilepsy Res. 2018;139:92–101. doi: 10.1016/j.eplepsyres.2017.11.016. [DOI] [PubMed] [Google Scholar]
- 6.Kang SH, Scheffer A, Ou Z, Li J, Scaglia F, Belmont J, et al. Identification of proximal 1p36 deletions using array-CGH: a possible new syndrome. Clin Genet. 2007;72:329–38. doi: 10.1111/j.1399-0004.2007.00876.x. [DOI] [PubMed] [Google Scholar]
- 7.Battaglia A, Hoyme HE, Dallapiccola B, Zackai E, Hudgins L, McDonald-McGinn D, et al. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics. 2008;121:404–10. doi: 10.1542/peds.2007-0929. [DOI] [PubMed] [Google Scholar]
- 8.Redon R, Rio M, Gregory SG, Cooper RA, Fiegler H, Sanlaville D, et al. Tiling path resolution mapping of constitutional 1p36 deletions by array-CGH: contiguous gene deletion or “deletion with positional effect” syndrome? J Med Genet. 2005;42:1666–71.. doi: 10.1136/jmg.2004.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenfeld JA, Crolla JA, Tomkins S, Bader P, Morrow B, Gorski J, et al. Refinement of causative genes in monosomy 1p36 through clinical and molecular cytogenetic characterization of small interstitial deletions. Am J Med Genet A. 2010;152A:1951–9. doi: 10.1002/ajmg.a.33516. [DOI] [PubMed] [Google Scholar]
- 10.Buck A, du Souich C, Boerkoel CF. Minimal genotype–phenotype correlation for small deletions within distal 1p36. Am J Med Genet A. 2011;155A:3164–9. doi: 10.1002/ajmg.a.34333. [DOI] [PubMed] [Google Scholar]
- 11.Heilstedt HA, Burgess DL, Anderson AE, Chedrawi A, Tharp B, Lee O, et al. Loss of the potassium channel beta-subunit gene, KCNAB2, is associated with epilepsy in patients with 1p36 deletion syndrome. Epilepsia. 2001;42:1103–11. doi: 10.1046/j.1528-1157.2001.08801.x. [DOI] [PubMed] [Google Scholar]
- 12.Colmenares C, Heilstedt HA, Shaffer LG, Schwartz S, Berk M, Murray JC, et al. Loss of the SKI proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Ski-/- mice. Nat Genet. 2002;30:106–9. doi: 10.1038/ng770. [DOI] [PubMed] [Google Scholar]
- 13.Gajecka M, Yu W, Ballif BC, Glotzbach CD, Bailey KA, Shaw CA, et al. Delineation of mechanisms and regions of dosage imbalance in complex rearrangements of 1p36 leads to a putative gene for regulation of cranial suture closure. Eur J Hum Genet. 2005;13:139–49. doi: 10.1038/sj.ejhg.5201302. [DOI] [PubMed] [Google Scholar]
- 14.Windpassinger C, Kroisel PM, Wagner K, Petek E. The human gamma-aminobutyric acid A receptor delta (GABRD) gene: molecular characterisation and tissue-specific expression. Gene. 2002;292:25–31. doi: 10.1016/S0378-1119(02)00649-2. [DOI] [PubMed] [Google Scholar]
- 15.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet. 2004;13S1:R57–64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 16.Heilstedt HA, Shapira SK, Gregg AR, Shaffer LG. Molecular and clinical characterization of a patient with duplication of 1p36.3 and metopic synostosis. Clin Genet. 1999;56:123–8. doi: 10.1034/j.1399-0004.1999.560205.x. [DOI] [PubMed] [Google Scholar]
- 17.Giannikou K, Fryssira H, Oikonomakis V, Syrmou A, Kosma K, Tzetis M, et al. Further delineation of novel 1p36 rearrangements by array-CGH analysis: narrowing the breakpoints and clarifying the “extended” phenotype. Gene. 2012;506:360–8. doi: 10.1016/j.gene.2012.06.060. [DOI] [PubMed] [Google Scholar]
- 18.Khan J, Toews H, Wang JC, Arredondo J, Provias J, Göhring G, et al. Acute lymphoblastic leukemia in a patient with constitutional chromosome 1pter-p36.31 duplication and 1q43-qter deletion. J Pediatr Hematol Oncol. 2012;34:217–21. doi: 10.1097/MPH.0b013e31823321e5. [DOI] [PubMed] [Google Scholar]
- 19.Tonk VS, Wilson GN, Yatsenko SA, Stankiewicz P, Lupski JR, Schutt RC, et al. Molecular cytogenetic characterization of a familial der(1)del(1)(p36.33)dup(1)(p36.33p36.22) with variable phenotype. Am J Med Genet A. 2005;139A:136–40. doi: 10.1002/ajmg.a.30958. [DOI] [PubMed] [Google Scholar]
- 20.Dobyns WB, Mirzaa G, Christian SL, Petras K, Roseberry J, Clark GD, et al. Consistent chromosome abnormalities identify novel polymicrogyria loci in 1p36.3, 2p16.1-p23.1, 4q21.21-q22.1, 6q26-q27, and 21q2. Am J Med Genet A. 2008;146A:1637–54. doi: 10.1002/ajmg.a.32293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saito Y, Kubota M, Kurosawa K, Ichihashi I, Kaneko Y, Hattori A, et al. Polymicrogyria and infantile spasms in a patient with 1p36 deletion syndrome. Brain Dev. 2011;33:437–41. doi: 10.1016/j.braindev.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Baart EB, Martini E, Van Opstal D. Screening for aneuploidies of ten different chromosomes in two rounds of FISH: a short and reliable protocol. Prenat Diagn. 2004;24:955–61. doi: 10.1002/pd.1052. [DOI] [PubMed] [Google Scholar]
- 23.Andrieu D, Watrin F, Niinobe M, Yoshikawa K, Muscatelli F, Fernandez PA. Expression of the Prader-Willi gene Necdin during mouse nervous system development correlates with neuronal diffenreciation and P75NTR expression. Gene Expr Patterns. 2003;5:761–5. doi: 10.1016/S1567-133X(03)00138-8. [DOI] [PubMed] [Google Scholar]
- 24.El Waly B, Buhler E, Haddad MR, Villard L. Nhej1 deficiency causes abnormal developement of the cerebral cortex. Mol Neurobiol. 2014 doi: 10.1007/s12035-014-8919-y. [DOI] [PubMed] [Google Scholar]
- 25.Stutterd CA, Leventer RJ. Polymicrogyria: a common and heterogeneous malformation of cortical development. Am J Med Genet. 2014;166C:227–39. doi: 10.1002/ajmg.c.31399. [DOI] [PubMed] [Google Scholar]
- 26.Kyukwang K, Junghyun E, Inkyung J. Characterization of Structural Variations in the Context of 3D Chromatin Structure. Mol Cells. 2019;42:512–22. doi: 10.14348/molcells.2019.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, et al. Mammalian Gene Collection Program Team. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci USA. 2002;99:16899–903. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapira SK, McCaskill C, Northrup H, Spikes AS, Elder FF, Sutton VR, et al. Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome. Am J Hum Genet. 1997;61:642–50. doi: 10.1086/515520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faivre L, Morichon-Delvallez N, Viot G, Martinovic J, Pinson MP, Aubry JP, et al. Prenatal detection of a 1p36 deletion in a fetus with multiple malformations and a review of the literature. Prenat Diagn. 1999;19:49–53. doi: 10.1002/(SICI)1097-0223(199901)19:1<49::AID-PD450>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 30.Slavotinek A, Shaffer LG, Shapira SK. Monosomy 1p36. J Med Genet. 1999;36:657–63. [PMC free article] [PubMed] [Google Scholar]
- 31.Shiba N, Daza RA, Shaffer LG, Barkovich AJ, Dobyns WB, Hevner RF. Neuropathology of brain and spinal malformations in a case of monosomy 1p36. Acta Neuropathol Commun. 2013;1:45. doi: 10.1186/2051-5960-1-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilhais-Neto GC, Maruhashi M, Smith KT, Vasseur-Cognet M, Peterson AS, Workman JL, et al. Rere controls retinoic acid signalling and somite bilateral symmetry. Nature. 2010;463:953–7. doi: 10.1038/nature08763. [DOI] [PubMed] [Google Scholar]
- 33.Wang L, Rajan H, Pitman JL, McKeown M, Tsai CC. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 2006;20:525–30. doi: 10.1101/gad.1393506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yanagisawa H, Bundo M, Miyashita T, Okamura-Oho Y, Tadokoro K, Tokunaga K, et al. Protein binding of a DRPLA family through arginine-glutamic acid dipeptide repeats is enhanced by extended polyglutamine. Hum Mol Genet. 2000;9:1433–42. doi: 10.1093/hmg/9.9.1433. [DOI] [PubMed] [Google Scholar]
- 35.Zoltewicz JS, Stewart NJ, Leung R, Peterson AS. Atrophin 2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis. Development. 2004;13:3–14. doi: 10.1242/dev.00908. [DOI] [PubMed] [Google Scholar]
- 36.Kim BJ, Zaveri HP, Shchelochkov OA, Yu Z, Hernández-García A, Seymour ML, et al. An allelic series of mice reveals a role for RERE in the development of multiple organs affected in chromosome 1p36 deletions. PLoS ONE. 2013;8:e57460. doi: 10.1371/journal.pone.0057460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim BJ, Scott DA. Mouse model reveals the role of RERE in cerebellar foliation and the migration and maturation of Purkinje cells. PLoS ONE. 2014;9:e87518. doi: 10.1371/journal.pone.0087518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fregeau B, Kim BJ, Hernández-García A, Jordan VK, Cho MT, Schnur RE, et al. De novo mutations of RERE cause a genetic syndrome with features that overlap those associated with proximal 1p36 deletions. Am J Hum Genet. 2016;98,:963–70. doi: 10.1016/j.ajhg.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordan VK, Fregeau B, Ge X, Giordano J, Wapner RJ, Balci T, et al. Genotype-phenotype correlations in individuals with pathogenic RERE variants. Hum Mutat. 2018;39:666–75. doi: 10.1002/humu.23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joseph J, Cruz-Sánchez FF, Carreras J. Enolase activity and isoenzyme distribution in human brain regions and tumors. J Neurochem. 1996;66:2484–90. doi: 10.1046/j.1471-4159.1996.66062484.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.