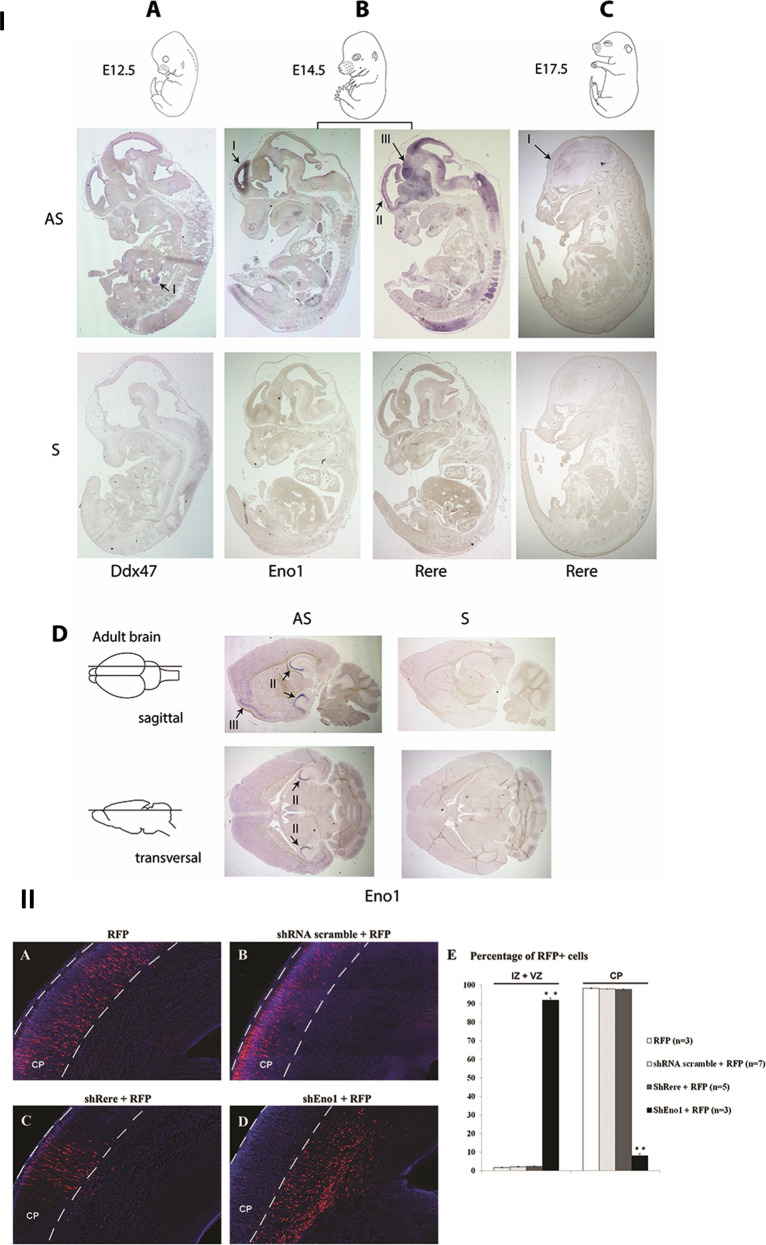
Fig. 4. In situ hybridization and in utero electroporation.
I. Expression of Eno1, Rere, and Ddx47 in mouse development using in situ hybridization. a–c Sagittal sections of mouse embryo at E12.5 (a), E14.5 (b), and E17.5 (c). d Sagittal and transverse sections of adult brains. Eno1 was strongly expressed in the inner ventricular side of the neopallial cortex at E14.5 (b–I), in the CA2 and CA3 fields of hippocampus and in the cerebral cortex in the adult brain (d-II and d-III, respectively). Rere expression was detected in central nervous system and strongly in the neopallial cortex (outer side) (b-II) and the midbrain (b-III) at E14.5. At E17.5, Rere expression was also detected in the neopallial cortex but at a lower level (c-I). A faint expression of Ddx47 transcript was detected in the urogenital region at E12.5 (a-I). II. Coronal sections of rat brain at E20, five days after electroporation. The presence of RFP alone (a), of a scramble shRNA (b), or shRere-2 (c) does not disturb neuronal migration. The presence of shEno1-1 severely impairs neuronal migration (d). Cells were counted in the different regions of the developing brain: intermediate (IZ) or ventricular (VZ) zones, or the cortical plate (CP) and plotted for comparison (E). Results are reported as mean ± standard error of the mean (SEM), and Student t test was used to test statistical significance, p value = 0.0008 for the significative difference in IZ and VZ and p value = 0.0009 for the significative difference in CP. A p value < 0.001 was considered to be statistically significant. **p < 0.001.