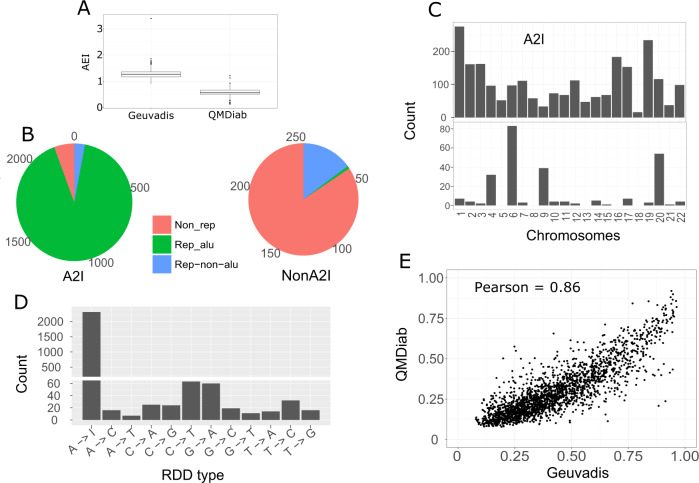
Fig. 2. Characteristics of detected A2I and non-A2I.
a Alu editing index [35] shows a higher RNAe activity in Geuvadis than in QMDiab. b Pie chart describing the percentage of A2I and nonA2I RDDs falling in Alu repetitive regions, non-Alu repetitive regions, and nonrepetitive regions. c Distribution of A2I and non-A2I RDDs across the human chromosomes. A2I RDDs are distributed evenly among regions with sequencing reads, while non-A2I RDDs are enriched in specific genomic regions (d) Types of RDD detected. (e). Correlation between overall (mean) RNAe level of 2,119 A2I RNAe sites between two independent datasets.