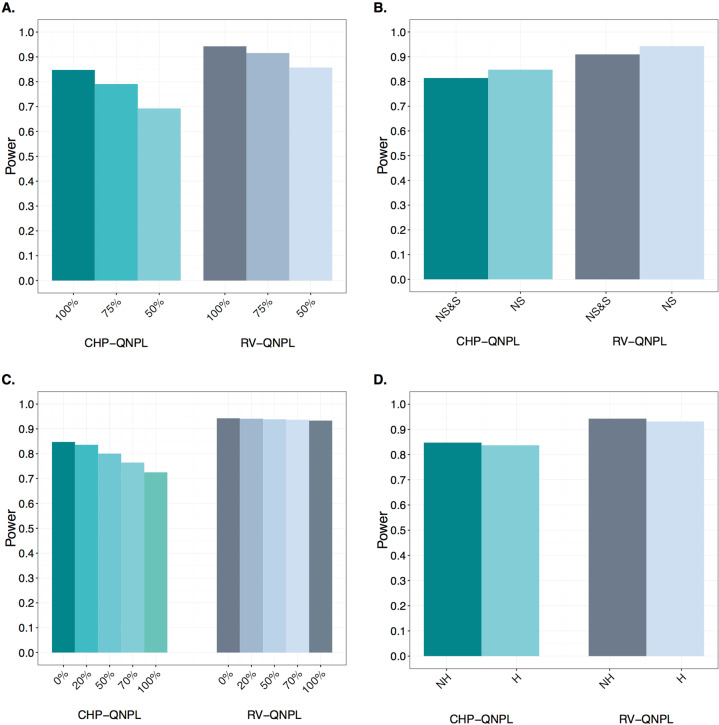
Fig. 2. Exome-wide power for RV-QNPL and CHP-QNPL in extended pedigrees.
One-hundred extended pedigrees with simulated QT and genotype data were analyzed with 100%, 75%, and 50% of the RVs being causal and the remaining non-causal (a); only causal nonsynonymous (NS) RVs as well as causal nonsynonymous (NS) and non-causal synonymous (S) RVs (b); 0%, 20%, 50%, 70% and 100% of founders missing both their genotype and phenotype data (c); and under linkage homogeneity [No heterogeneity (NH)], i.e., 100 linked families as well as with locus heterogeneity (H), i.e., 100 linked and 50 unlinked families (d).