Abstract
The opportunistic Pseudomonas aeruginosa virulence controlled by quorum sensing (QS) also identified as, cell-cell communication. QS system is organized by the LasI-LasR and the RhlI-RhlR components. Provided that QS tends to perform a key role in virulence gene expression and host defence function, QS inhibitors have been proposed as potential antipseudomonal therapies. Sub-inhibitory concentrations (sub-MIC) of antibiotics, although having biostatic effect on bacteria, but can interfere with bacterial QS system and virulence. This research aimed to examine the impact of sub-MIC of azithromycin, imipenem, cefepime and piperacillin/tazobactam on the QS-dependent virulence including pyocyanin and biofilm production, haemolysin, protease and DNase in P. aeruginosa wildtype and mutant strains; transcriptional-regulator (ΔLasR), autoinducer synthesis protein (ΔLasI), transcriptional-regulator (ΔRhlR), protease precursor (ΔLasA) and double regulators mutants (ΔLasR/RhlR). The growth of all strains showed similar pattern, however, in presence of antibiotics significant growth variation was observed among mutant strains when compared to wild type strain. Antimicrobial activity tested by agar diffusion method of all antibiotics on all strains were used to compare the zones of therapeutic and sub-MIC doses showing a significant difference in the inhibition zone. QS-dependant virulence as biofilm, pyocyanin, protease, haemolysin and DNase production showed significant variation on all strains compared to wild type in response to antibiotics used at sub-MIC doses. In conclusion well known antibiotics can be used in sub-MIC doses to decrease the virulence of P. aeruginosa in addition to overcoming the major side effect of the high doses and the occurrence of resistance.
Keywords: Pseudomonas aeruginosa, Quorum sensing, QS-dependant virulence, Sub-inhibitory concentrations, LasI-LasR, RhlI-RhlR
1. Introduction
Quorum sensing has involved in regulation of Pseudomonas aeruginosa virulence elements such as elastase, protease, haemolysin, pyocyanin, swimming and twitching motilities, biofilm production and resistance to oxidative-stress, which is a type of intercellular signalling in bacteria, that produce signalling molecules and autoinducers. Once the quorum comes to a critical concentration it activate genes that encode for the virulence factors in the bacteria (Stover et al., 2000, El-Mowafy et al., 2017). P. aeruginosa have a main pair of quorum sensing; lasI/lasR and the rhlI/rhlR systems (Whiteley et al., 1999, Venturi, 2006). The lasI/lasR and the rhlI/rhlR systems are connected and las system employs a positive regulator over the rhl system stimulating both rhlI and rhlR transcription (Pesci et al., 1997, Duan and Surette, 2007). lasI pr oduce 3-oxodecanoyl homoserine lactone (C12 HSL) that binds to lasR receptor, while rhlI produces butanoyl homoserine lactone (C4 HSL) that is recognized by rhlR receptor. Although both lasI/lasR and rhlI/rhlR are co-regulated, the LasI/LasR system is dominant (Whiteley et al., 1999; Venturi, 2006; Pesci et al., 1997). Hence, the inhibition of these systems will hinder the pathogenicity of P. aeruginosa, and it might be utilized for the treatment or prevention of infections caused by P. aeruginosa (Smith and Iglewski, 2003, El-Mowafy et al., 2017). Many studies have examined infectious behaviour of P. aeruginosa using strains which have omissions of one or more of the QS-related genes, and wildtype strains by a burnt-mouse model, a murine design of acute-pneumonia, and a rat design of chronic lung-infection (O’Loughlin et al., 2013). They found that mutually the las and the rhl QS systems are crucial for P. aeruginosa to induces both acute and chronic lung infections, causing morbidity and mortality. Furthermore, a study on cystic fibrosis patients colonized with P. aeruginosa showed that the QS genes transcription level correlate with those of QS-regulated genes which means that their expression during infection was regulated by QS. In addition, when they analyse patient’s sputum they could directly measure AHLs. Another in vivo experiment demonstrates that several inflammatory cytokines and chemokines can be stimulated by 3O-C12-HSL, also 3O-C12-HSL could acts as an immunosuppressors (Smith and Iglewski, 2003). Another study was performed to identify the QS controlling genes by utilizing DNA-microarrays, P. aeruginosa wild type strain and autoinducers deficient strain (lasI, rhlI) were used for expressing analysis. They found a high percentage (>10%) of genome associated with QS regulation, which reflects that QS has a large influence on cellular behaviour (Wagner et al., 2003).
Another study was performed in 2017, investigating the effect of sub-inhibitory concentrations of β-lactam antibiotics; ceftazidime, cefepime and imipenem on Quorum sensing signals using reporter strain assay and on the virulence factors (elastase, protease, pyocyanin and haemolysin) of P. aeruginosa, and the results showed significant eradication of the QS signals 3OH-C12-HSL and C4-HSL, and suppressed their virulence factors up to 1/20 MIC (El-Mowafy et al., 2017, Smith and Iglewski, 2003). Moreover, on 2017 a study was performed to investigate the effect of sub-MIC concentrations of pyridoxal lactohydrazone (32 and 8 μg/mL) against P. aeruginosa Quorum sensing related virulence factor (Heidari et al., 2017). They performed molecular docking for assessing the effect of pyridoxal lactohydrazone with the LasR receptor, and it showed that pyridoxal lactohydrazone has potential to inhibit the LasR protein. It demonstrated significant inhibition of other virulence factors involving motility, alginate and pyocyanin formation and susceptibility to H2O2 (Heidari et al., 2017). Another study screened 12 different antibiotics for their QS inhibition ability on P. aeruginosa, using a simple plate diffusion screening based on QS inhibitor selector 1 (QSIS1), they found that no activity or low levels of QSI with spectinamycin, chloramphenicol, tobramycin, griseofulvin, gentamicin, kanamycin, tetracycline, piperacillin, and streptomycin (Skindersoe et al., 2008b). On the other hand, azithromycin, ciprofloxacin, and ceftazidime showed high QSI activities. Also, they compared P. aeruginosa QS mutants with wild type and found that the tolerance to antibiotic was decreased [10]. All these findings may help to target the QS by therapeutics to eradicate the P. aeruginosa infection (Singh et al., 2017, Skindersoe et al., 2008b). Additional study validating the effect of a frequently used antibiotic, doxycycline on QS with sensor strains of Chromobacterium violaceum and P. aeruginosa PAO1. Doxycycline at Sub-MICs significantly compact QS-related virulence in C. violaceum, and P. aeruginosa PAO1. The results of this study highlight the multiple activities of doxycycline against QS-associated virulence factors and its potential to diminish virulence of P. aeruginosa (Husain and Ahmad, 2013).
The first line for treating bacterial infections is antibiotics, but because of the high emergence of resistance, it is necessary to look for new therapeutic approaches to overcome this global problem. One of the important features of these new therapeutic approaches is to target the virulence factors rather than introducing a stress on the bacteria that could lead to acquiring multiple resistance (Skindersoe et al., 2008b, Livermore, 2002). In this study, we aim to investigate the effect of Azithromycin, Meropenem, Cefepime and Piperacillin/tazobactam, at sub-inhibitory dose on QS dependent-virulence factors as a potential treatment strategy for pseudomonal infection.
2. Materials and methods
2.1. Bacterial strains, chemicals, and growth media
All Pseudomonas aeruginosa strains used in this study and their source are described in (Table1). Pseudomonas aeruginosa strains PAOI (WT), PA1430 (ΔlasR), PA1432(ΔlasI), PA1817(ΔlasA), and PA3477 (ΔrhlR) were purchased from the Manoil laboratory at the University of Washington Seattle and ΔlasR ΔrhlR (DA6) knockouts mutant strains obtained from Oregon State University Corvallis, OR, United States (Asfahl and Schuster, 2018). Reference strain P. aeruginosa PAO1 and mutant strains were routinely cultured in Luria-Bertani (LB) agar or broth (Lab M limited, United Kingdom) or Cetrimide agar (Lab M limited, United Kingdom), supplemented with antibiotics as required. All strains were grown aerobically at 37 °C. All antibiotics (azithromycin, imipenem, cefepime and piperacillin/tazobactam) were gifted from King Khalid University Hospital, Saudi Arabia and dissolved in deionized water. The concentration of used antibiotics were as follow: inhibitory concentration (MIC90) and sub-inhibitory concentration (MIC12.5). Azithromycin MIC90 = 64 mg/L, MIC12.5 = 8 mg/L. Meropenem MIC90 = 2 mg/L, MIC12.5 = 0.25 mg/L. Piperacillin/Tazobactam MIC90 = 16 mg/L, MIC12.5 = 2 mg/L. Cefepime MIC90 = 8 mg/L, MIC12.5 = 1 mg/L.
Table 1.
Description of Pseudomonas aeruginosa strains used in this study.
| Strain name | Abbreviation | Description |
|---|---|---|
| Parent strain | PAOI | PAOI wild type |
| PA1430 | ΔlasR | transcriptional regulator LasR |
| PA1432 | ΔlasI | autoinducer synthesis protein LasI |
| PA3477 | ΔrhlR | transcriptional regulator RhlR |
| PA1871 | ΔlasA | LasA protease precursor |
| DA6 | ΔlasR ΔrhlR double mutant | ΔlasR ΔrhlR knockouts double-nul mutant |
2.2. Spot test
First, isolated colony of all tested strains was cultured in 5 mL of LB broth and incubated at 37 °C with continues shaking at 200 rpm overnight. On the day of the test, we prepared 7 mL of 0.7% agarose with sterile water was prepared and 50 µl of bacterial broth grew at range of 0.4 to 0.6 was added to agarose to reach 0.5 McFarland standard then the mixture gently poured into ceramide plate. We made 20 ceramide plates, each 5 plates were used for single strain, and each plate of those 5, was tested for single antibiotic using 3 concentrations MIC90, MIC12.5 and MIC25, then the plates were incubated at 37 °C for 24 h. The experiment repeated three times.
2.3. Growth curve
All P. aeruginosa strains were cultured in 5 mL LB broth with selected antibiotic marker “tetracycline” except for the wildtype strain, then incubated at 37 °C with continues shaking at 200 rpm overnight. On the day of the test the optical density of all bacterial broth was measured and normalized at OD600 nm with LB broth, then LB broth containing antibiotic concentrations of (MIC90, MIC12.5) were prepared and 100 µl was added to Bioscreen C 100 well microtiter plates (Labsystems Oy, Helsinki, Finland). To designated wells, 20 µl of the 0.5 McFarland bacterial culture was added. Controls wells that contain media only and media with bacteria, were also included and plates were placed in Bioscreen C and optical density of bacterial growth was recoded at 600 nm for 24 h.
2.4. Antibiotic susceptibility determining
Minimum inhibitory concentrations (MIC) and sub-MIC for all used antibiotics in this study were determined according to the EUCAST guidelines by the microdilution broth method as described previously (Wiegand et al., 2008, Microbiology and Diseases, 2000).
2.5. Preparation of P. aeruginosa culture, supernatants and cell extracts
Overnight cultures of all P. aeruginosa strains used in this study were standardized to OD600 0.5 and 1:50 diluted in 10 mL LB with or without antibiotics (sub-MIC 12.5). After 24 h incubation at 37 °C, the cell pellets and supernatant were collected and filter-sterilized. For cells extract cell P. aeruginosa cells pellet were harvested by centrifugation (Beckman microfuge; 10 min) at 4 °C, then cells were suspended in lysis buffer and kept at −20 °C for further analysis (Kessler et al., 1998). All experiments were performed three times as triplicates.
2.6. Measurements of pyocyanin
Regarding pyocyanin detection, 3 mL of chloroform was added to 7.5 mL supernatant of P. aeruginosa culture and vortexed. The chloroform phase was kept after centrifugation (5000 rpm, 5 min), and mixed with 1.5 mL HCl (0.2 M). The absorbance of the pink HCl layer was measured at 520 nm with the EnVision Multilabel Reader after vortexing and centrifugation previously (Aleanizy et al., 2018, Krishnan et al., 2012). 0.2 M HCl was used as a negative control in the measurement of absorbance.
2.7. Protease activity by agar plate assay
The proteolytic activity of all strains was carried out as described (Kiratisin et al., 2002) with some modification. Briefly, grown culture and supernatant of all stains were streaked on agar plates containing 5% skim milk and the plates were incubated overnight at 30 °C for 48hs then placed at 4 °C for another 24hs. Proteolysis was visible as clear zones around the growing bacteria. The zones of clearance were observed and compared for all strains.
2.8. Haemolysis assay
For Haemolytic activity an overnight cultures of all P. aeruginosa strains used in this study were standardized to OD600 0.5 and 1:50 diluted in 10 mL LB with or without antibiotics (sub-MIC 12.5). Strains were streaked on blood agar plates incubated overnight at 37 °C for 24 and 48 hs. The Haemolytic activity was observed on plates as transparency around the colonies (Burnside et al., 2010).
2.9. Dnase assay
The test for the detection of exoenzyme Dnase by P. aeruginosa PAO1 and mutant strains was carried out utilizing Dnase test agar (Oxoid). In brief, the strains were streaked on a Dnase test agar plate and incubated at 37 °C for 24 h according to manufacturer’s instructions. The Dnase test agar composed of tryptose, DNA and NaCl. The production of Dnase resulted in the hydrolysis of DNA in the media. Following incubation, the plates were flooded with 1 N HCl to observe any clearing or hydrolysis of DNA by the bacterium (Kumar et al., 2019).
2.10. Biofilm assay
Biofilm detection was performed as previously described with some modification (Shih and Huang, 2002, Bukhari and Aleanizy, 2020). Briefly, overnight grown strains in LB broth at 37 °C, following day the cultures were diluted in LB media to 107cfu/ml and dispensed in 96-well microtiter plate (Thomas science), antibiotic at sub-inhibitory concentration, MIC12.5 (meropenem, azithromycin, cefepime and pipecoline/tazobactam), and 100 µl will be transferred to the 96-well plate. For each strain, 15-wells will be inoculated (100 µl of bacterial strain will be added), as the antibiotics will be added in triplicate for each strain and three wells as negative control to check biofilm inhibition, the plates were then incubated at 37 °C for 24 h. Cell suspension was decanted, and the plates subsequently were washed 2 times with 0.9% NaCl and inverted to dry at room temperature for 1 h. Afterward, 150 µl of crystal violet solution (CV; Prolab Diagnostics) was added to the wells and was allowed to stain for 15 min. After staining, CV was discarded, and the wells were washed three times with 0.9% NaCl. The attached CV was then solubilized by adding 200 µl of ethanol-acetone (80:20 v/v). Eventually, the absorbance of CV was read at 595 nm using microplate reader (BioTek). All experiments were carried out in triplicate three independent times.
2.11. Statistical analysis
Differences between experimental groups were evaluated using unpaired two-tailed Student's t-test. A -value of 0.05 considered significant. All experiments were performed in triplicates for three independent times. Results were expressed as averages SD.
3. Results
3.1. Growth pattern of Pseudomonas QS mutants in the presence of antibiotics
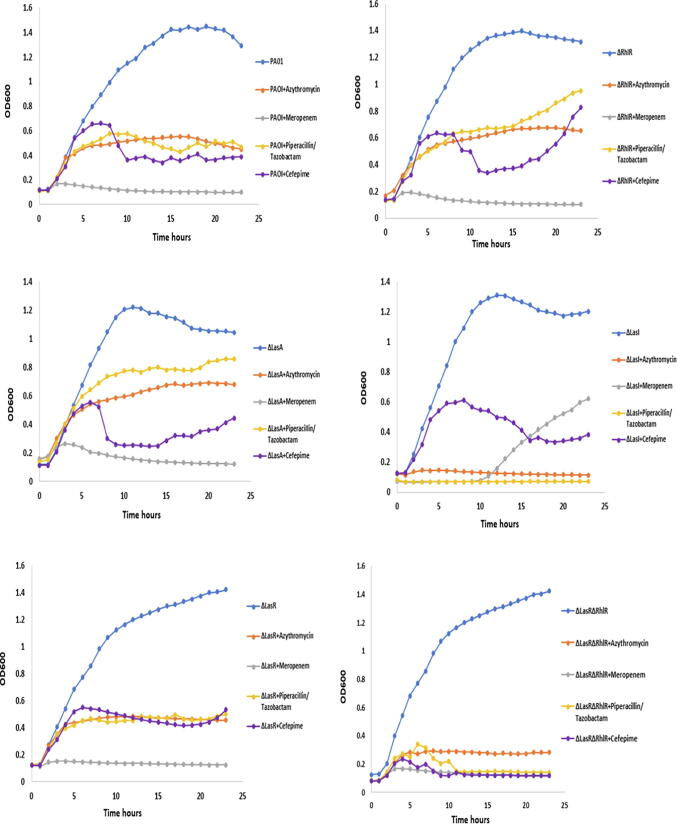
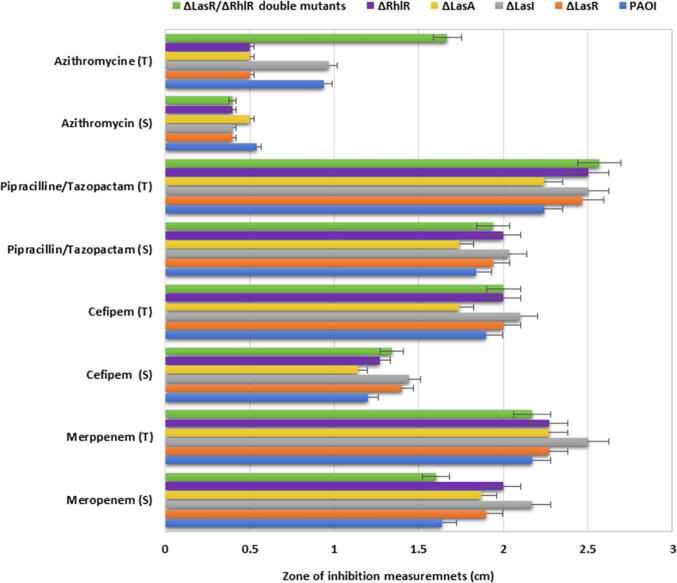
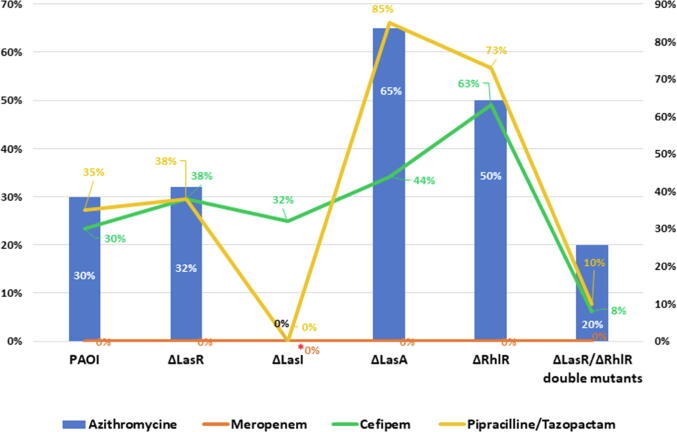
The effect of different antibiotics at sub-inhibitory dose on QS systems of P. aeruginosa have been studied previously (Skindersoe et al., 2008a), however, other antibiotics including piperacillin/tazobactam, and cefepime used in this study were not yet investigated. The effect of these antibiotics at sub-inhibitory dose was investigated using wild type PA01 and different pseudomonal QS mutants PA1430 (ΔLasR), PA1432 (ΔlasI), PA3477 (ΔrhlR), PA1871 (ΔlasA) and D6A (ΔLasR/ΔrhlR double mutants). The selected sub-inhibitory concentration of antibiotics was chosen according to the least effective inhibition on PA01 according to EUCAST guidelines. Therefore, MIC12.5 was selected for azithromycin, meropenem and piperacillin/tazobactam, and MIC25 for cefepime for all subsequent experiments. First the growth of all strains was evaluated, and the results showed that all strains exhibited similar growth pattern (Fig. 1A). In the presence of azithromycin at sub-MIC concentration, the growth of all strains was reduced by pattern displaying 70%, 68%,35%, 50% and 80.3% reduction in growth of Wt, ΔLasR, ΔLasA, ΔRhlR and ΔlasR/ΔRhlR double mutants respectively, except ΔlasI, where the growth was completely inhibited (Fig. 1B). This was furtherly confirmed by biostatic effect observed when measuring the inhibition zone of therapeutic and sub-inhibitory dose of azithromycin on all strains (Fig. 2A, Fig. 2B). In case of using piperacillin/tazobactam at sub-MIC, growth of wild type, ΔLasR and ΔlasR/ΔRhlR double mutants reduced by 65%, 62%, and 90%, respectively, while ΔrhlR and ΔlasA growth reduced by 27% and 15%, respectively, and complete inhibition of ΔlasI growth (Fig. 1). Cefepime was also used in the current study, however, MIC25 concentration was used and its effect shown in Fig. 1. Cefepime at sub-MIC reduced the growth by 70%, 68.3%, 62.2%, 56%, 37% and 92% of wild type, ΔlasI, ΔLasR, ΔlasA, ΔrhlR and ΔlasR/ΔRhlR double mutants respectively. Meropenem is an antipseudomonal antibiotic and utilized in this study at sub-MIC for comparison with other classes of antibiotics and demonstrated complete killing of all strains except ΔlasI, which restored their viability after 10 hr of inhibition (Fig. 1). To furtherly visualize the antimicrobial activity of antibiotics, agar diffusion method was used, and results illustrated in Fig. 2A, Fig. 2B were the therapeutic and sub-MIC doses of all antibiotics compared and showed significant differences with (p ≤ 0.01). To validate the significance of the sub-MIC effect on all strains, the percentage of cell survival were calculated as shown in (Fig. 3).
Fig. 1.
The effect of selected antibiotics at sub-inhibitory dose was investigated using wild type PA01 and different pseudomonal QS mutants PA1430 (ΔLasR), PA1432 (ΔlasI), PA3477 (ΔrhlR), PA1871 (ΔlasA) and ΔLasR/ΔrhlR double mutants. All experiment performed three times in triplicate.
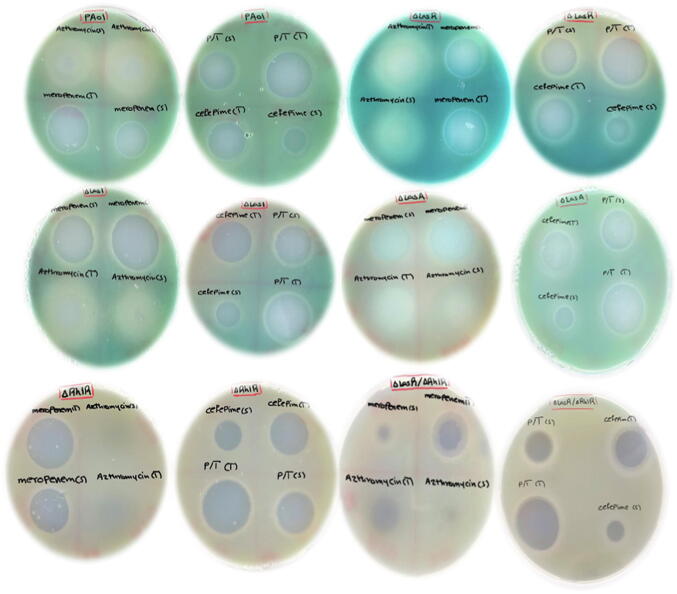
Fig. 2A.
Measuring antibiotic activity using plate diffusion method. Each antibiotic assayed as therapeutic and sub-MIC doses to compare. The therapeutic and sub-MIC doses of all antibiotics compared and showed significant differences with (p ≤ 0.01). All experiment performed three times in triplicate.
Fig. 2B.
Comparing the therapeutic and sub-MIC doses of antibiotic activity using plate diffusion method showing significant differences with (p ≤ 0.01). All experiment performed three times in triplicate.
Fig. 3.
Percentage of cell survival of Pseudomonas aeruginosa and its QS mutants in presence of sub-MIC antibiotics. All experiment performed three times in triplicate.
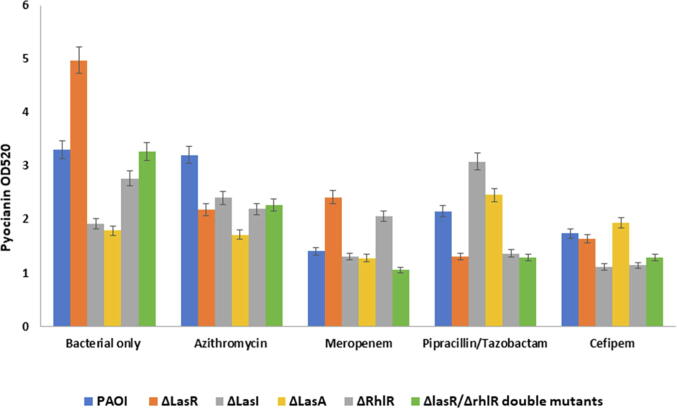
3.2. Pyocyanin production of P. Aeruginosa and its QS mutants in response to the antibiotics
The effect of antibiotics at sub-MIC dose against the pyocyanin production in P. aeruginosa wildtype and mutated strains was evaluated. As illustrated in (Fig. 4), the pyocyanin production decreased significantly (p ≤ 0.001) in wild type in response to meropenem, piperacillin/tazobactam, and cefepime but not with azithromycin (p ≥ 0.08). In ΔLasR and ΔrhlR mutants, all antibiotics significantly (p ≤ 0.0001) reduced pyocyanin production. In contrast, significant enhance in pyocyanin production was observed in ΔlasA in response to azithromycin, piperacillin/tazobactam, and cefepime but not meropenem (Fig. 4). With ΔlasI, azithromycin and piperacillin/tazobactam increased pyocyanin production significantly while meropenem and cefepime reduced pyocyanin production. In the ΔLasR/ΔrhlR double mutants pyocyanin production reduced but was not significant. Pyocyanin production of different strains was also visualized and was obvious on plates when measuring the antibiotic zone of inhibition on cetrimide plate as increased in greenish pigmentation (Fig. 2A).
Fig. 4.
Effect of pyocyanin production on Pseudomonas aeruginosa and its QS mutants. All experiment performed three times in triplicate.
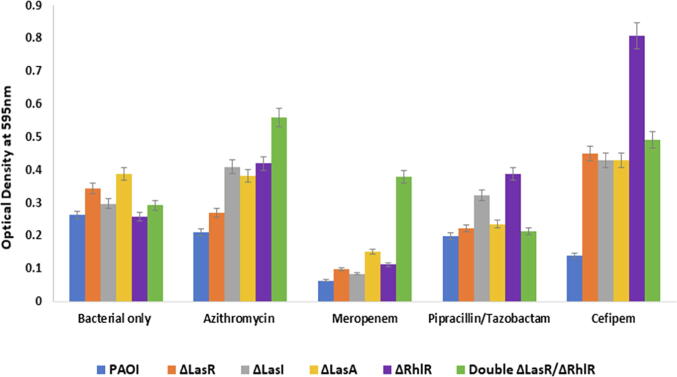
3.3. Biofilm formation of Pseudomonas QS mutants in response to the antibiotics
The effect of antibiotics at MIC12.5 on biofilm production was reduced significantly (p ≤ 0.02) with meropenem in ΔlasR and ΔlasI stains compared to control strains. However, none significant reduction on the biofilm in ΔlasA (p ≤ 0.07) and ΔrhlR (p ≤ 0.2) except the double mutants ΔlasR/ΔrhlRthat showed increase by 29.3% (Fig. 4). With azithromycin, biofilm formation reduced in WT, Δlasr and ΔlasA none significantly (with a p value ≥ 0.2), however, an increase in biofilm production by 37.8%, 62.4% and 91.2% with ΔlasI, ΔrhlR and double mutants ΔlasR/ΔrhlR, respectively. Biofilm production was enhanced by piperacillin/tazobactam in ΔlasI and ΔrhlR by 8.3% and 50%, respectively. While in WT, ΔlasR, ΔlasA and double mutants ΔlasR/ΔrhlR, piperacillin/tazobactam reduced biofilm formation by 24.3%, 35.4%, 40% and 27% (Fig. 4). Although cefepime reduced biofilm production in WT, an increase in biofilm formation in Δlasr, ΔlasA, ΔlasI and double mutants ΔlasR/ΔrhlR by 31%, 44.7%, 11.4% and 67.8% was observed and markedly induced in ΔrhlR by 212.3% (p ≤ 0.04) (Fig. 5).
Fig. 5.
Effect of antibiotics on the biofilm formation of Pseudomonas aeruginosa and its QS mutants. All experiment performed three times in triplicate.
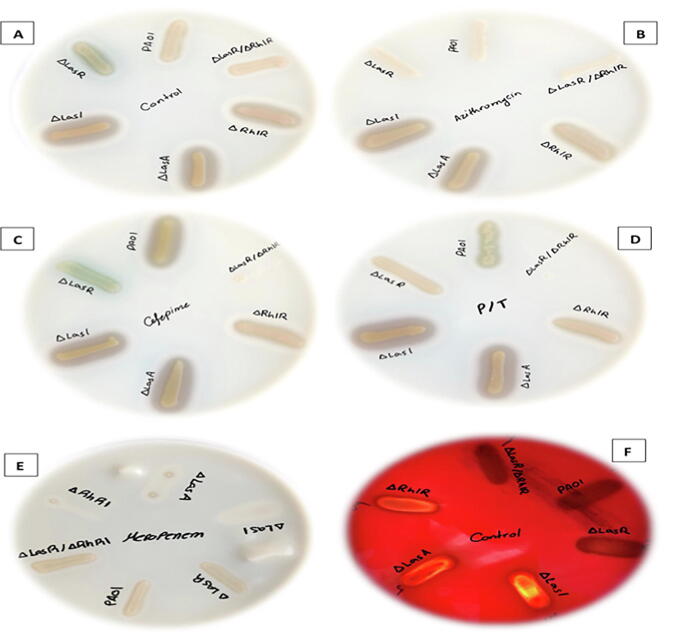
3.4. Protease, haemolysin and DNase production in Pseudomonas aeruginosa QS mutants in response to the sub-MIC antibiotics
Other virulence factors including protease, haemolysin and DNase production were evaluated. As observed in Fig. 6, protease productions displayed different pattern with used antibiotics among tested strains. In control milk plates, WT, ΔLasR and ΔlasR/ΔrhlR double mutants showed similar proteolytic zone, whereas ΔlasI, ΔrhlR and ΔlasA showed similar zone but were more than WT and other strains (Fig. 6A). Adding Azithromycin to milk plate (Fig. 6B) cause abolishing of protease production in WT, ΔLasR and ΔlasR/ΔrhlR double mutants, however, ΔlasI, ΔrhlR and ΔlasA showed similar pattern compared to control plate. On milk agar supplemented with meropenem (Fig. 6E), very weak growth on the plate for ΔlasI, ΔlasA, ΔrhlR but WT, ΔLasR, and ΔlasR/ΔrhlR double mutants grow with no proteolytic zone to compare. With Piperacillin/tazobactam (Fig. 6D), all strains showed same zone of proteolysis compared to control except ΔlasR/ΔrhlR double mutants that was abolished completely. With cefepime (Fig. 6C), WT have shown more proteolytic activity compare to control, while ΔlasR/ΔrhlR double mutants showed abolished activity and ΔLasR produced less zone compared to control. Regarding haemolysin (Fig. 6F) and DNase activities (results not shown), there were no significant changes in the haemolysis and DNase pattern of bacteria in the presence of antibiotics at sub-inhibitory level when compared with bacteria cultured without antibiotics. For haemolytic activity, WT, ΔLasR and ΔlasR/ΔrhlR double mutants showed partial haemolysis whereas ΔlasI, ΔrhlR and ΔlasA showed complete haemolysis.
Fig. 6.
Visualizing growth of Pseudomonas aeruginosa and its QS mutant’s strains. (panels A-E) protease activity measured as zone around each strain either control plate or with the addition of antibiotics, (panel F) haemolytic activity of each strains on sheep blood agar control plate only. (panel A) control plates no antibiotics added. All plates grown in triplicate, repeated independently three times.
4. Discussion
Quorum sensing has been a potential goal for the treatment of Pseudomonas aeruginosa infection where several studies have investigated its part in controlling the virulence of Pseudomonas aeruginosa (Davies et al., 1998, Wilder et al., 2011). With the advance of antibiotic resistance, the exploration for new approaches past conventional antibiotics becomes a key aim of health research; however, in this study we searched within conventional drugs previously classified for their antibiotic actions and screened if they possess properties that interfere with communication and, theoretically, have quorum sensing inhibitory activity. Previous studies demonstrated the effect of azithromycin on the QS-regulated virulence by showing improvement in clinical outcome in CF model indicating that screening the well-known antibiotics at a sub-MIC dose may serve as a maintenance or protective therapy (Hentzer et al., 2003, Yang et al., 2012). Another study screened twelve antibiotics and found three of them with QSI activity (Skindersoe et al., 2008b).
Although meropenem used at sub-MIC dose its activity is maintained in wild type, LasR, LasA, LasI and RhIR strains showing complete killing and reduction in proteolytic activity and biofilm production, except LasR/RhIR that showed 10% increase in biofilm secretion. The loss of single QS gene components had not affect meropenem activity, except LasI mutants were the strain restored their growth after 10 hrs which might be due to stimulation of other QS systems; however, the exact mechanism is not investigated. Notably, the loss of both QS regulatory components has led to increase the stain sensitivity to meropenem indicating a role of these genes in the activity of meropenem. The observed decrease in pyocyanin and abolished proteolytic activity of pseudomonal wildtype and its QS mutated strains in company of meropenem corroborates previous studies results (Fothergill et al., 2007, Fuse et al., 2013). In the current study, meropenem at sub-MIC decreased biofilm formation significantly in wild type, LasR, LasA, LasI and RhIR strains and inversely induced biofilm formation in LasR/RhIR strain. This variable effect of meropenem on biofilm capability indicated that its effect is dependent on strain type. It might also suggest that meropenem may act on other genetic determinants involve in biofilm formation other than LasR/RhIR genes (Maura et al., 2016). PT have a minimal killing activity at sub-MIC dose on wild type and LasR, LasA, and RhIR. Loss of LasA led to bacterial growth by 85% when exposed to PT and increase pyocyanin and biofilm production, although loss of RhIR led to bacterial growth by 73% and increases in biofilm production it decreases pyocyanin production. The loss of LasI gene have cause the strains to be sensitive to PT, as PT is known to have the ability to bind to protein and mutating LasI autoinducer synthesis protein might affect the mechanism by which PT kills the cells. The loss of LasA protease precursor and RhlR transcriptional regulator individually increased the resistance of the cells to PT. However, knocking out both genes (LasR and RhLR) increased strain sensitivity to PT and reduced biofilm, pyocyanin formation and abolished proteolytic activity. The reduction in biofilm formation in response to PT at sub-MIC was reported for PA01 strain previously which was in agreement with our study (Fonseca et al., 2004). However, according to our knowledge other QS strains virulence phenotypes were not investigated in response to PT previously. Of note, deletion of LasR reduced biofilm formation in both LasR and LasR/RhLR strain.
Azithromycin at sub-MIC dose has minimal killing activity on wild type, LasR and LasR/RhlR, and complete killing activity on LasI; however, the loss of LasA protease precursor gene have led the strain to be more resistance to azithromycin were cells grows by 65% compared to WT. It is well known that azithromycin mechanism of action of preventing bacteria from growing by interfering with their protein synthesis, it seems that the protease activity have indirect synergistic effect on azithromycin.
Cefepime at sub-MIC dose has killing activity on WT, LasR, LasI and LAsR/RhlR but less killing activity on strain LasA and RhlR stains. The observed effect of cefepime on WT showed similar results indicated in a previous study using third generation cephalosporin, ceftazidime (Husain et al., 2016). The loss of RhlR transcriptional regulator gene and LasA protease precursor gene might interfere with cefepime activity that impedes the final transpeptidation step of peptidoglycan synthesis. Cefepime reduced biofilm formation in wild type but induced it in other strains, particularly strain lacking RhlR gene.
Of note, all antibiotics used in this study decreased biofilm formation in wild type PA01; however, they induced biofilm formation in QS mutant strains in different pattern. This induction pattern in response to exposure to sub MIC dose of antibiotics was reported for other bacterial species including methicillin-resistant S. aureus (MRSA) (Ng et al., 2014), Enterococcus faecalis (Yu et al., 2018), and S. epidermidis (He et al., 2016). This signifies the importance of studying molecular genetics of clinical isolates.
In Conclusion
QS perform a main role in the expression of virulence and communication with host defence. The studied antibiotics can be used at sub-MICs in combination with other antipseudomonal therapies to reduce the virulence, help to overcome bacterial resistance, and avoiding the major side effects of the high doses.
Author Contributions
F.S.A. conceived and designed the experiments, write and revised the paper, and led the project and performed lab experiments and contributed reagents and materials. F.Y.A. performed experiments, analysed the data, and wrote the initial paper. N.A performed lab experiments, analysed the data. R.A. performed lab experiments and analysed the data. E.K.E. performed lab experiments analysed the data. A.A. contributed materials, analysed the data and revised the paper. T.A.A. analysed the data and revised the paper. H.A. analysed the data and revised the paper. All authors confirmed the final manuscript.
Declaration of Competing Interest
The authors report no conflicts of interest in this work.
Acknowledgments
The authors are very grateful to the Deanship of Scientific Research and Research Centre, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia. Also the authors would like to thank Dr. Martin Schuster for his gift of previously constructed strain of double mutant.
Footnotes
Peer review under responsibility of King Saud University.
References
- Aleanizy F.S., Alqahtani F.Y., Shazly G., Alfaraj R., Alsarra I., Alshamsan A., Abdulhady H.G. Measurement and evaluation of the effects of pH gradients on the antimicrobial and antivirulence activities of chitosan nanoparticles in Pseudomonas aeruginosa. Saudi Pharmaceut. J. 2018;26:79–83. doi: 10.1016/j.jsps.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asfahl K.L., Schuster M. Additive Effects of Quorum Sensing Anti-Activators on Pseudomonas aeruginosa Virulence Traits and Transcriptome. Front. Microbiol. 2018;8 doi: 10.3389/fmicb.2017.02654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukhari S.I., Aleanizy F.S. Association of OprF mutant and disturbance of biofilm and pyocyanin virulence in pseudomonas aeruginosa. Saudi Pharmaceut. J. 2020;28(2):196–200. doi: 10.1016/j.jsps.2019.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnside, K., Lembo, A., De Los Reyes, M., Iliuk, A., Binhtran, N.-T., Connelly, J. E., Lin, W.-J., Schmidt, B. Z., Richardson, A.R., Fang, F.C., 2010. Regulation of hemolysin expression and virulence of Staphylococcus aureus by a serine/threonine kinase and phosphatase. PloS One, 5, e11071. [DOI] [PMC free article] [PubMed]
- Davies D.G., Parsek M.R., Pearson J.P., Iglewski B.H., Costerton J.W., Greenberg E.P. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science. 1998;280:295–298. doi: 10.1126/science.280.5361.295. [DOI] [PubMed] [Google Scholar]
- Duan K., Surette M.G. Environmental Regulation of Pseudomonas aeruginosa PAO1 Las and Rhl Quorum-Sensing Systems. J. Bacteriol. 2007;189:4827–4836. doi: 10.1128/JB.00043-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mowafy S.A., el Galil K.H.A., Habib E.-S.E., Shaaban M.I. Quorum sensing inhibitory activity of sub-inhibitory concentrations of β-lactams. African Health Sci. 2017;17:199–207. doi: 10.4314/ahs.v17i1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca A., Extremina C., Fonseca A., Sousa J. Effect of subinhibitory concentration of piperacillin/tazobactam on Pseudomonas aeruginosa. J. Med. Microbiol. 2004;53:903–910. doi: 10.1099/jmm.0.45637-0. [DOI] [PubMed] [Google Scholar]
- Fothergill J.L., Panagea S., Hart C.A., Walshaw M.J., Pitt T.L., Winstanley C. Widespread pyocyanin over-production among isolates of a cystic fibrosis epidemic strain. BMC Microbiol. 2007;7:45. doi: 10.1186/1471-2180-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse K., Fujimura S., Kikuchi T., Gomi K., Iida Y., Nukiwa T., Watanabe A. Reduction of virulence factor pyocyanin production in multidrug-resistant Pseudomonas aeruginosa. J. Infect. Chemother. 2013;19:82–88. doi: 10.1007/s10156-012-0457-9. [DOI] [PubMed] [Google Scholar]
- He H.-J., Sun F.-J., Wang Q., Liu Y., Xiong L.-R., Xia P.-Y. Erythromycin resistance features and biofilm formation affected by subinhibitory erythromycin in clinical isolates of Staphylococcus epidermidis. J. Microbiol. Immunol. Infect. 2016;49:33–40. doi: 10.1016/j.jmii.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Heidari A., Noshiranzadeh N., Haghi F., Bikas R. Inhibition of quorum sensing related virulence factors of Pseudomonas aeruginosa by pyridoxal lactohydrazone. Microb. Pathog. 2017;112:103–110. doi: 10.1016/j.micpath.2017.09.043. [DOI] [PubMed] [Google Scholar]
- Hentzer M., Wu H., Andersen J.B., Riedel K., Rasmussen T.B., Bagge N., Kumar N., Schembri M.A., Song Z., Kristoffersen P. Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J. 2003;22:3803–3815. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain F.M., Ahmad I. Doxycycline interferes with quorum sensing-mediated virulence factors and biofilm formation in Gram-negative bacteria. World J. Microbiol. Biotechnol. 2013;29:949–957. doi: 10.1007/s11274-013-1252-1. [DOI] [PubMed] [Google Scholar]
- Husain Fohad Mabood, Ahmad Iqbal, Baig Mohammad Hassan, Khan Mohammad Shavez, Khan Mohd Shahnawaz, Hassan Iftekhar, Al-Shabib Nasser Abdulatif. Broad-spectrum inhibition of AHL-regulated virulence factors and biofilms by sub-inhibitory concentrations of ceftazidime. RSC Adv. 2016;6(33):27952–27962. doi: 10.1039/C6RA02704K. [DOI] [Google Scholar]
- Kessler E., Safrin M., Gustin J.K., Ohman D.E. Elastase and the LasA protease of Pseudomonas aeruginosa are secreted with their propeptides. J. Biol. Chem. 1998;273:30225–30231. doi: 10.1074/jbc.273.46.30225. [DOI] [PubMed] [Google Scholar]
- Kiratisin P., Tucker K.D., Passador L. LasR, a transcriptional activator of Pseudomonas aeruginosa virulence genes, functions as a multimer. J. Bacteriol. 2002;184:4912–4919. doi: 10.1128/JB.184.17.4912-4919.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan T., Yin W.-F., Chan K.-G. Inhibition of Quorum Sensing-Controlled Virulence Factor Production in Pseudomonas aeruginosa PAO1 by Ayurveda Spice Clove (Syzygium Aromaticum) Bud Extract. Sensors. 2012;12(4):4016–4030. doi: 10.3390/s120404016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Sheemal Shanista, Penesyan Anahit, Elbourne Liam Davin Hunt, Gillings Michael R., Paulsen Ian T. Catabolism of Nucleic Acids by a Cystic Fibrosis Pseudomonas aeruginosa Isolate: An Adaptive Pathway to Cystic Fibrosis Sputum Environment. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01199.s003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livermore D.M. Multiple mechanisms of antimicrobial resistance in Pseudomonas aeruginosa: our worst nightmare? Clin. Infect. Dis. 2002;34(5):634–640. doi: 10.1086/338782. [DOI] [PubMed] [Google Scholar]
- Maura D., Hazan R., Kitao T., Ballok A.E., Rahme L.G. Evidence for direct control of virulence and defense gene circuits by the Pseudomonas aeruginosa quorum sensing regulator, MvfR. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep34083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Microbiology, E.C.F.A.S.T.O.T.E.S.O.C. & DISEASES, I. 2000. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Clin. Microbiol. Infect., 6, 509-515. [DOI] [PubMed]
- Ng Mandy, Epstein Samuel B., Callahan Mary T., Piotrowski Brian O., Simon Gary L., Roberts Afsoon D., Keiser John F., Kaplan Jeffrey B. Induction of MRSA Biofilm by Low-Dose β-Lactam Antibiotics: Specificity, Prevalence and Dose-Response Effects. Dose-Response. 2014;12(1) doi: 10.2203/dose-response.13-021.Kaplan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Loughlin C.T., Miller L.C., Siryaporn A., Drescher K., Semmelhack M.F., Bassler B.L. A quorum-sensing inhibitor blocks Pseudomonas aeruginosa virulence and biofilm formation. Proc. Natl. Acad. Sci. 2013;110:17981–17986. doi: 10.1073/pnas.1316981110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesci E.C., Pearson J.P., Seed P.C., Iglewski B.H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih P.-C., Huang C.-T. Effects of quorum-sensing deficiency on Pseudomonas aeruginosa biofilm formation and antibiotic resistance. J. Antimicrob. Chemother. 2002;49:309–314. doi: 10.1093/jac/49.2.309. [DOI] [PubMed] [Google Scholar]
- Singh V.K., Mishra A., Jha B. Anti-quorum sensing and anti-biofilm activity of Delftia tsuruhatensis extract by attenuating the quorum sensing-controlled virulence factor production in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017;7:337. doi: 10.3389/fcimb.2017.00337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skindersoe M.E., Alhede M., Phipps R., Yang L., Jensen P.O., Rasmussen T.B., Bjarnsholt T., Tolker-Nielsen T., Hoiby N., Givskov M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008;52:3648–3663. doi: 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skindersoe M.E., Alhede M., Phipps R., Yang L., Jensen P.O., Rasmussen T.B., Bjarnsholt T., Tolker-Nielsen T., Høiby N., Givskov M. Effects of antibiotics on quorum sensing in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2008;52:3648–3663. doi: 10.1128/AAC.01230-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.S., Iglewski B.H. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Investig. 2003;112:1460–1465. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C., Pham X., Erwin A., Mizoguchi S., Warrener P., Hickey M., Brinkman F., Hufnagle W., Kowalik D., Lagrou M. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406:959. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- Venturi V. Regulation of quorum sensing in Pseudomonas. FEMS Microbiol. Rev. 2006;30(2):274–291. doi: 10.1111/j.1574-6976.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- Wagner V.E., Bushnell D., Passador L., Brooks A.I., Iglewski B.H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley M., Lee K.M., Greenberg E. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 1999;96:13904–13909. doi: 10.1073/pnas.96.24.13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Wilder Cara N., Diggle Stephen P., Schuster Martin. Cooperation and cheating in Pseudomonas aeruginosa: the roles of the las, rhl and pqs quorum-sensing systems. ISME J. 2011;5(8):1332–1343. doi: 10.1038/ismej.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-X., Xu Z.-H., Zhang Y.-Q., Tian J., Weng L.-X., Wang L.-H. A new quorum-sensing inhibitor attenuates virulence and decreases antibiotic resistance in Pseudomonas aeruginosa. J. Microbiol. 2012;50:987–993. doi: 10.1007/s12275-012-2149-7. [DOI] [PubMed] [Google Scholar]
- Yu W., Hallinen K.M., od K.B. Interplay between antibiotic efficacy and drug-induced lysis underlies enhanced biofilm formation at subinhibitory drug concentrations. Antimicrob. Agents Chemother. 2018;62:e01603–e01617. doi: 10.1128/AAC.01603-17. [DOI] [PMC free article] [PubMed] [Google Scholar]