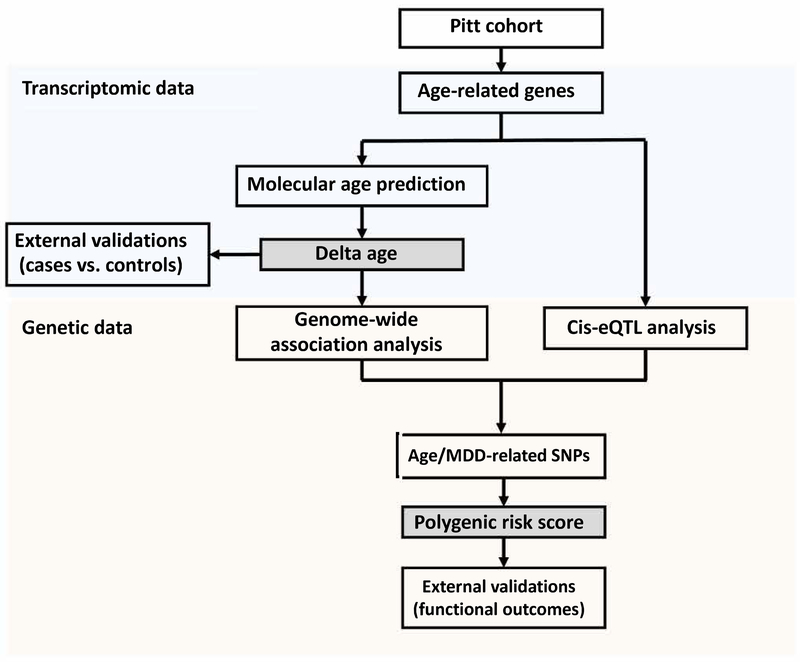
Figure 1. Study flow chart.
Firstly, transcriptomic data from the Pitt cohort (BA47 and BA11) were used to identify age-related genes. A molecular age prediction model was constructed by using top age-related genes. “Delta age” was defined as the difference between molecular and chronological ages, which served as a proxy measure of molecular brain aging. We then tested the robustness of the findings in external cohorts. Secondly, genetic data from the European subjects in same cohorts in was used to identify SNPs associated with brain aging. Two approaches (Genome-wide association and Cis-eQTL analyses) were used to narrow down the potential SNP lists. Polygenic risk scores were defined for each subject as a cumulative score measuring how much genetic risk a subject carries toward premature brain aging. Finally, our proposed risk scores were validated against clinical diagnoses and related functional outcomes.