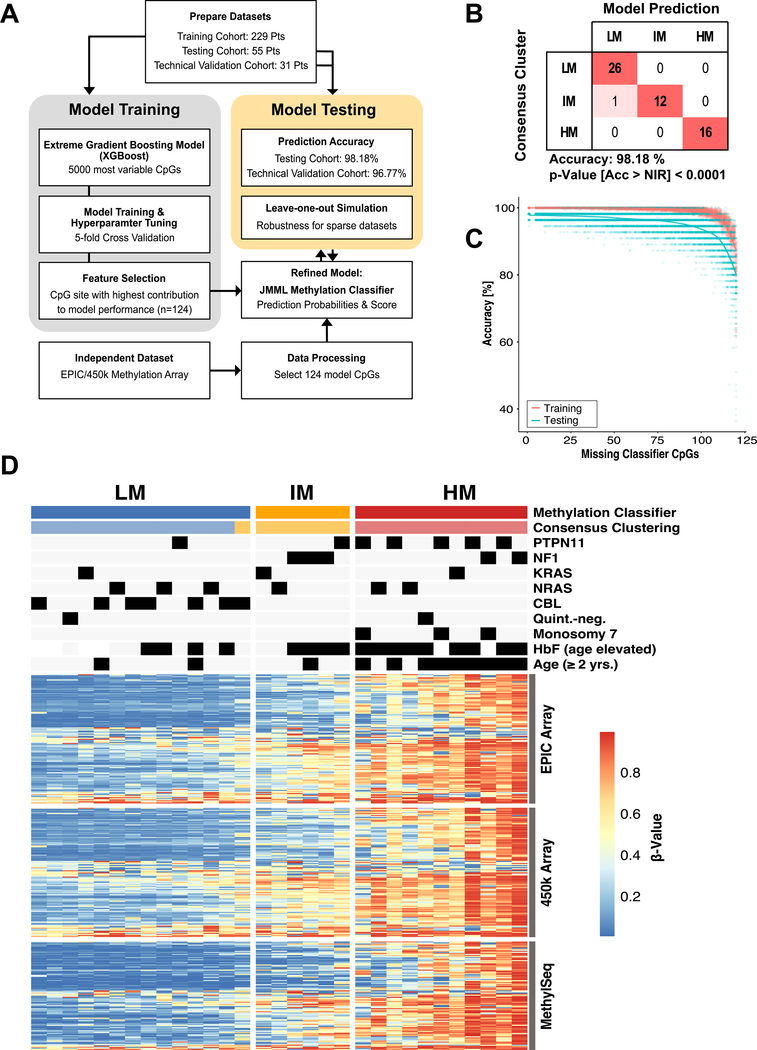
Figure 3 |. Development of a DNA methylation classifier for JMML.
(A) The meta-analysis cohort was randomly split into a training (n = 229) and a testing cohort (n=55). A machine learning classifier was built based on the 5000 mvCpGs, which were covered by 450k and EPIC methylation arrays and trained on the training cohort. The fractional contribution of each CpG to the overall model performance was calculated and the CpGs with the highest gain were selected (n=124). This refined JMML methylation classifier was trained and the model performance evaluated on the testing cohort. Additionally, 31 patients were re-analyzed on EPIC arrays to test the model accuracy across different technical assays (“technical validation cohort”). (B) Confusion table for prediction of the testing cohort using the refined JMML methylation classifier (Acc, accuracy; NIR, no-information rate). (C) The JMML methylation classifier was re-trained with subsets of the 124 model CpGs, by repeatedly leaving out between 5 and 120 CpG sites. The model accuracy of these sparse models for predicting the training or testing cohort was determined. (D) Heatmap showing the DNA methylation beta-values for the 124 model CpG sites (rows) on 450k and EPIC methylation arrays for patients of the technical validation cohort (n=31; columns). Additionally, 104/124 CpGs assessed by the MethylSeq assay are shown. The predictions by the methylation classifier model are compared to the consensus clustering.