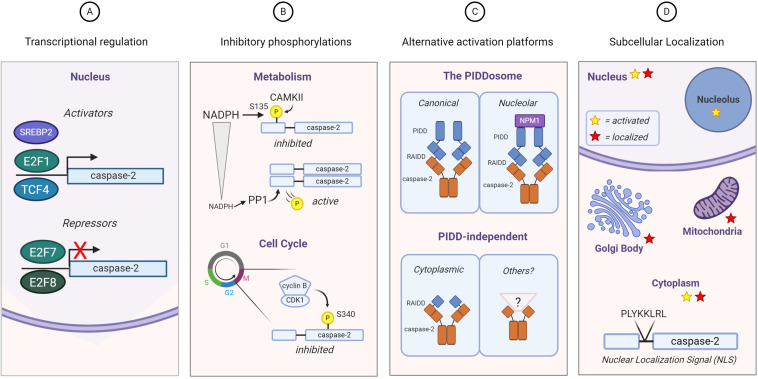
FIGURE 3.
Regulation of caspase-2 expression, activity, and localization. Substrate selection by caspase-2 could be directed by (A) transcriptional regulation: E2F family transcription factors both activate (E2F1) and repress (E2F7 and E2F8) caspase-2 transcription. TCF4 and SREBP2 activate caspase-2 transcription; (B) phosphorylation: Caspase-2 is phosphorylated on multiple residues. The known phosphorylation events of caspase-2 result in its inhibition, which can be regulated by metabolism (top) and the cell cycle (bottom); (C) differential activation platform assembly: The canonical PIDDosome contains both PIDD1 and RAIDD. Caspase-2 can be activated by complexes lacking one or both of these elements. NPM1 is required for the nucleolar activation complex; (D) subcellular localization: Caspase-2 contains a classical nuclear localization signal and it has been found outside the nucleus associated with the Golgi body and mitochondria. Activation of caspase-2 has also been observed specifically in the nucleolus induced by DNA damage, as well as more generally throughout cytoplasm induced by heat shock and microtubule disruption. Red stars represent sites where caspase-2 is expressed and yellow stars represent sites where caspase-2 is activated.