Abstract
Objectives:
It remains difficult to characterize the source of pain in knee joints either using radiographs or magnetic resonance imaging (MRI). We sought to determine if advanced machine learning methods such as deep neural networks could distinguish knees with pain from those without it and identify the structural features that are associated with knee pain.
Methods:
We constructed a convolutional Siamese network to associate MRI scans obtained on subjects from the Osteoarthritis Initiative (OAI) with frequent unilateral knee pain comparing the knee with frequent pain to the contralateral knee without pain. The Siamese network architecture enabled pairwise learning of information from two-dimensional (2D) sagittal intermediate-weighted turbo spin echo slices obtained from similar locations on both knees. Class activation mapping (CAM) was utilized to create saliency maps, which highlighted the regions most associated with knee pain. The MRI scans and the CAMs of each subject were reviewed by an expert radiologist to identify the presence of abnormalities within the model-predicted regions of high association.
Results:
Using 10-fold cross validation, our model achieved an area under curve (AUC) value of 0.808. When individuals whose knee WOMAC pain scores were not discordant were excluded, model performance increased to 0.853. The radiologist review revealed that about 86% of the cases that were predicted correctly had effusion-synovitis within the regions that were most associated with pain.
Conclusions:
This study demonstrates a proof of principle that deep learning can be applied to assess knee pain from MRI scans.
Keywords: Knee, Osteoarthritis, Pain, Magnetic Resonance Imaging, Machine learning
INTRODUCTION
Osteoarthritis (OA) is the most common musculoskeletal disease and one of the leading causes of disability globally [1]. The most incapacitating manifestation of OA is pain, and painful OA is most common in the knee [2]. Severe OA-induced pain often leads to disability. Currently, there is no effective cure for advanced stage knee OA other than total joint replacement surgery. The occurrence of pain in knee joints with OA can be correlated with a variety of structural findings such as bone marrow lesions (BMLs), cartilage damage, synovitis, and effusion [3–7], as well as neuropathic mechanisms such as central sensitization and hyperalgesia [8–10]. Also, the frequency and severity of pain are self-reported and usually defined subjectively [8; 11].
Consequently, the correlation between pain and radiographic findings is weak and there has been little success in correlating OA-induced pain with a specific type and location of structural damage. One review found that the proportion of subjects with knee pain who have radiographic OA ranged from 15 to 76% [12]. Identifying the source and location of OA-induced pain could greatly benefit the design of targeted, individualized treatments to reduce symptoms and to limit disability [11]. Further, for those with knee pain as part of a widespread pain diathesis, determining the absence of pain-inducing knee pathology might aid in diagnosis.
MRI scans are capable of providing more detailed structural information about the knee joint than radiographs [13]. A systematic review of MRI measures found that knee pain may arise from BMLs, effusion and synovitis; however, the correlation between pain and MRI findings was inconsistent and moderate at best [14–16]. Further, since MRI findings associated with pain are often present in multiple locations in the knee and since many knees without pain also have MRI findings, the lesions associated with knee pain are hard to localize and identify. This limits treatment approaches that seek to target localized areas of the knee including surgeries such as unicondylar replacements and cryoneuroablation procedures, and hampers the development of targeted treatment strategies. Furthermore, rehabilitation strategies that focus on lessening the load to painful regions of the knee are limited. Hence, there is a need to develop a method to objectively and accurately associate MRI scans with knee pain. The first step of such an approach is to identify MRI findings that discriminate painful from nonpainful knees and then to determine the knee regions that are the likely sources of pain. In this paper, we pursue the first step, using deep learning to discriminate between painful and nonpainful knees. We begin to address the second step by identifying lesions identified in the first step as potential sources of pain.
Deep learning algorithms such as convolutional neural networks (CNN) can extract visual features, and one can utilize them for applications related to disease classification, segmentation and object detection [17; 18]. CNN model training is associated with learning a series of image filters through numerous layers of feed-forward neural networks. The filters are then projected on the original input image, and the image features that are most correlated with the outcome are extracted through the training process. Deep learning frameworks are increasingly being applied on MRI scans of different organ systems [19–23]. Recently, deep neural networks were applied on knee MRIs for segmenting different components of the knee [24–29], and for detection of ligament and meniscal tears [30].
The purpose of this study was first, to investigate the performance of a deep learning framework to differentiate painful knees from nonpainful ones and second, to identify the structural lesions that are most relevant to knee pain using MRI scans of both knees of individuals enrolled in the Osteoarthritis Initiative (OAI). Unilateral frequent knee pain was defined when an individual had pain, aching or stiffness for more than half of the days in a month in one knee and no pain in the contralateral knee. We used sagittal intermediate-weighted turbo spin echo (SAG-IW-TSE) sequence images that capture structural regions thought to be critical in generating knee pain, BMLs, synovitis, effusion and cartilage loss [4; 31–36], to train a convolutional Siamese network. We subsequently leveraged class activation mapping (CAM) to identify regions that were most associated with knee pain. An expert radiologist then independently reviewed the MRI scans and identified possible presence of knee abnormalities, which were then mapped with CAM-based findings.
METHODS
Detailed descriptions of the methods can be found in the supplement. Below we provided a brief summary.
Informed consent
Written informed consent was not required for this study because de-identified data is publicly available (Osteoarthritis Initiative: https://oai.nih.gov).
Study selection
We selected cases that had unilateral frequent knee pain who have undergone SAG-IW-TSE imaging from the baseline OAI dataset (n=4,796; Table 1) [37; 38]. A total of 1,505 subjects from them passed initial quality check (see below), and were used for construction of the deep learning model (Model A). Additionally, we excluded subjects with similar Western Ontario and McMaster Universities Osteoarthritis Index (WOMAC) pain scores between the left and right knee to construct another model. For this case, we selected a subset of subjects who had a WOMAC pain score difference greater than 2 between the knees. In total, 721 subjects met the criteria and 710 subjects were used for model construction after quality check (Model B).
Table 1:
Schematic of study selection. (Table 1a): In total, 1,606 subjects with unilateral frequent knee pain were selected from the OAI’s baseline study, consisting of 4,796 subjects. After image quality check, 1,505 subjects were selected to train the neural network (Model A). Subjects who had the contralateral difference in WOMAC pain score (≥ 3) were further selected, leading to 710 subjects which were then used to train another neural network (Model B). Stratified sampling was then used to divide the dataset in the ratio of 70:15:15, where 70% of the dataset was used for training, 15% for validation, and the remaining cases (n=107) for independent testing. The stratified sampling was performed to match for age, gender, and body mass index. These cases were then reviewed by the expert radiologist. (Tables 1b-1d): Baseline characteristics of the sub-group of individuals selected for model training (Models A & B), and for radiologist review.
| Number of subjects in OAI baseline dataset | Model A (n = 1505) | ||||
| n = 4796 | Parameter | Left Knee Pain | Right Knee Pain | p-value | |
| Cases with unilateral knee pain | Subjects | n = 734 | n = 771 | ||
| n = 1606 | Percent female | 57.9 | 55.9 | 0.700 | |
| Unilateral knee pain cases with selected after quality check (Model A) | Age (years) | 60.13 ± 9.18 | 61.24 ± 8.98 | 0.018 | |
| BMI (kg/m2) | 28.97 ± 4.88 | 28.18 ± 4.53 | 0.015 | ||
| n = 1505 | |||||
| Unilateral knee pain cases with contralateral WOMAC pain score difference (≥3) (Model B) | Model B (n = 710) | ||||
| Parameter | Left Knee Pain | Right Knee Pain | p-value | ||
| n = 107 | Subjects | n = 350 | n = 360 | ||
| Cases selected for radiologist review | Percent female | 59.4 | 60.8 | 0.854 | |
| n = 107 | Age (years) | 60.62 ± 9.39 | 61.21 ± 9.01 | 0.395 | |
| BMI (kg/m2) | 29.50 ± 4.96 | 28.94 ± 4.63 | 0.120 | ||
| MRI scan type | Cases for radiologist review (n = 107) | ||||
| SAG_IW_TSE_RIGHT, SAG_IW_TSE_LEFT | Parameter | Left Knee Pain | Right Knee Pain | p-value | |
| 2D MRI slice size | Subjects | n = 46 | n = 61 | ||
| 444×444 pixels | Percent female | 60.9 | 59.0 | 1.000 | |
| Age (years) | 60.83 ± 9.34 | 60.67 ± 8.90 | 0.932 | ||
| BMI (kg/m2) | 29.38 ± 5.30 | 29.01 ± 4.89 | 0.711 | ||
Image registration and quality check
We selected MRI slices with the most complete view of posterior cruciate ligament and indexed it as the center slice (Red colored box in Figure 1A), and indexed other slices relative to the center slice for each knee. We then performed Euclidean transformation to align the slice with respect to a previously selected template (Figure 1B). Images were then cropped and resized to 224×224 pixels, followed by selection of 11 adjacent slices on the lateral side of the center slice, 11 on the medial side of the center slice and the center slice for model training from each knee. We manually reviewed and discarded cases with missing data, abnormal misalignment within a slice, presence of a foreign object, and cases with sub-regions of high contrast (Figure 1C).
Figure 1: Image processing pipeline.
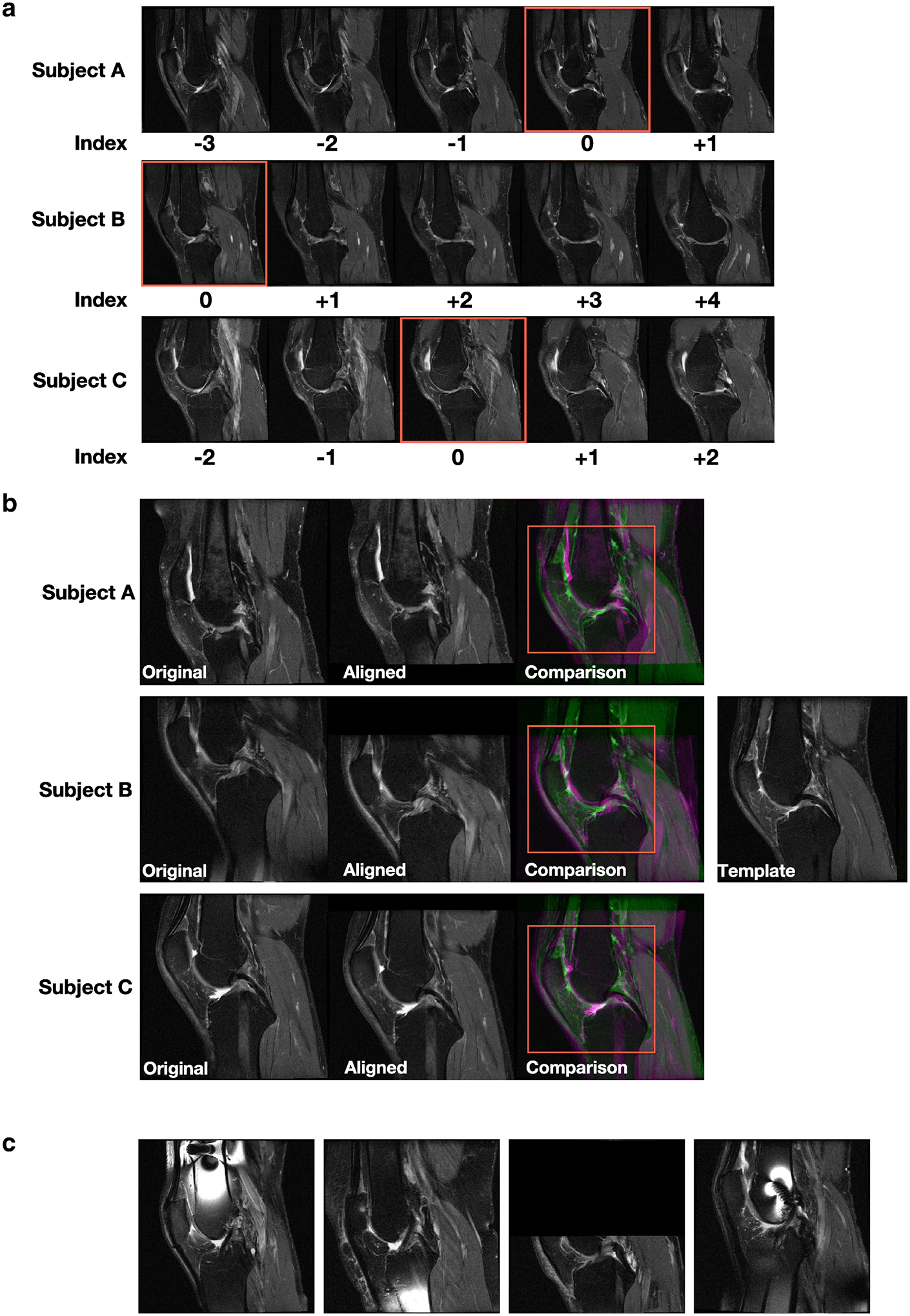
(A) For each subject’s knee joint, we manually examined all the MRI slices oriented in the sagittal view and selected the slice showing the posterior cruciate ligament (PCL) and indexed it as the center slice (red colored box). The remaining slices were indexed relative to the center slice for each knee. Two-dimensional MRI slices for three different subjects are shown. (B) For each 2D MRI slice of the knee for a subject, we performed linear registration to align the slice with respect to a template that was already selected after manual examination. Later, a region (red colored box) containing the center of the knee joint with dimensions 294×294 pixels was cropped for all registered slices and used for model training. Three cases from the baseline OAI dataset are shown. (C) Sample cases not used for model training due to the presence of various artifacts.
Neural network architecture
We developed a Siamese neural network architecture [39], such that a pair of MRI slices from the two knees, each extracted from a relatively similar location within each knee were learned together (Figure 2A). A series of convolutional operations, batch normalization, nonlinear activation, max pooling and average pooling were applied to predict knee pain, by solving a binary classification problem.
Figure 2: Convolutional Siamese network architecture.
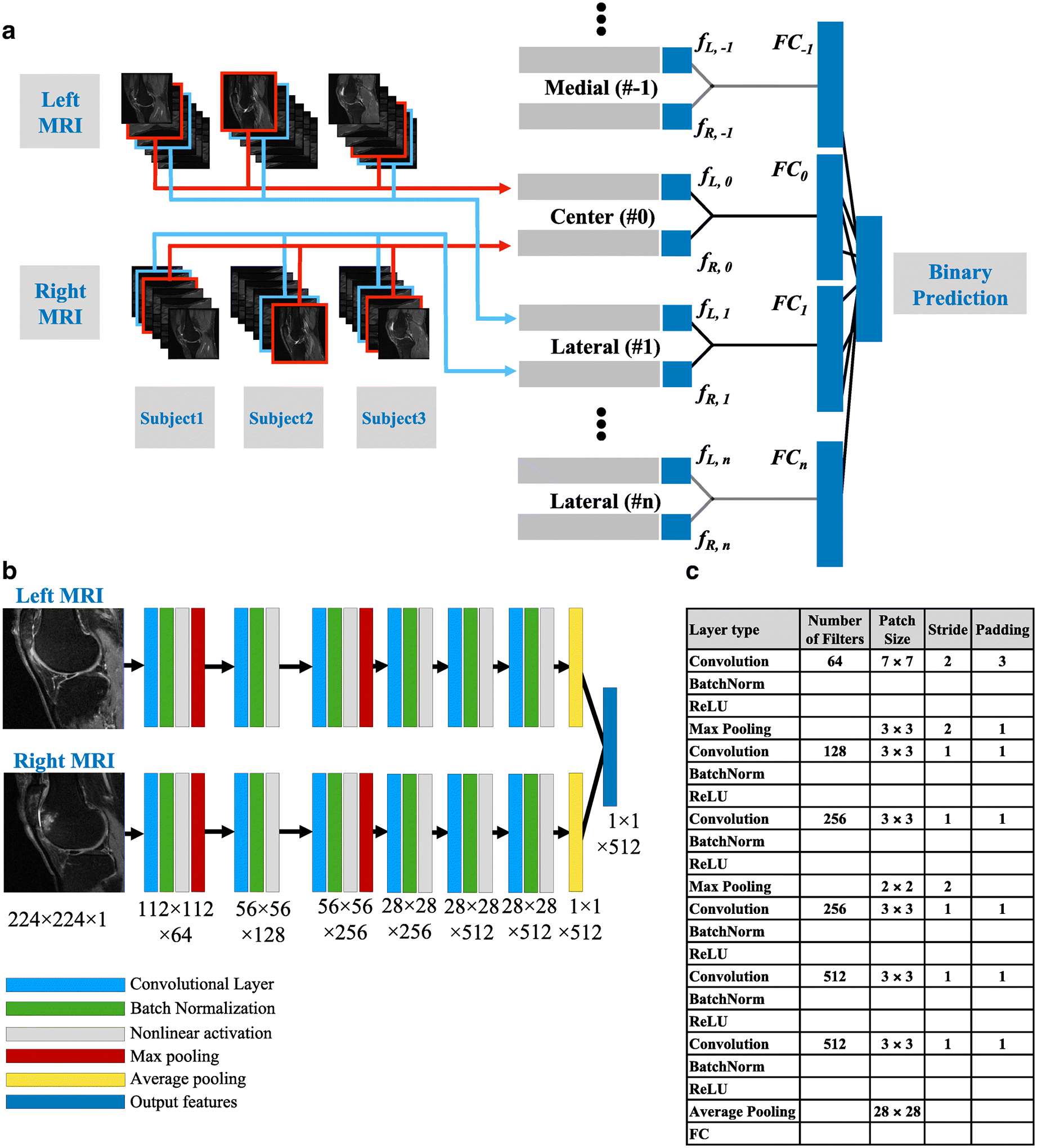
(A) After image pre-processing, the indexed MRI slices were fed into the corresponding Siamese network model. The outputs from all the 2D models were concatenated for the binary prediction task. (B) The 2D MR slices for the left and right knee were fed into the network simultaneously. Each neural network that is part of the Siamese architecture comprised of 6 convolutional layers. Black arrows represent connection between the images and the first layer, connections between the layers and the connection between the layer and the output. For layers 1 and 3, each convolutional operation was followed by batch normalization, nonlinear activation and max pooling, whereas for layers 2, 4, 5 and 6, each convolutional operation was followed by batch normalization and nonlinear activation. Only the first convolutional layer and the two max pooling layers had a stride of 2, whereas the other layers had a stride of 1. Consequently, the final convolutional layer had a high in-plane resolution with a dimension of 512×28×28. (C). Table presenting the details of the different layers within the neural network.
Class activation mapping
Class activation maps (CAMs) have the ability to localize the discriminative image regions from CNN models trained for classification without any prior locational knowledge [40]. To achieve this, we extracted the final feature maps of the left and right knee of the subject (fL,n and fR,n in Figure 2A, respectively), and multiplied them by weights of their respective fully-connected layer (FCn in Figure 2A), thus indicating the importance of each feature stored in the extracted maps. For each subject, we identified a CAM with the most pain-relevant regions by selecting the CAM with highest average value from the 23 CAMs generated from all the MRI slices.
Radiologist review
The dataset of 710 subjects (unilateral knee pain, between knee WOMAC pain difference ≥ 3) was divided into training, validation and testing sets in a 70:15:15 ratio (Table 2). A musculoskeletal radiologist with extensive experience in knee MRI interpretation reviewed CAMs of MRI of the last 15%, and identified the presence of abnormalities using the MRI scan. The radiologist then reviewed the model-derived CAMs and identified the specific lesion that was co-localized with the highlighted region in the CAM.
Table 2:
Data partitioning and modeling. Model A and Model B were constructed on the 1,505 subjects and 710 subjects, respectively. Performance of these models was evaluated after 10-fold cross validation. For radiologist review, we split the 710 cases in 70:15:15 ratio using stratified sampling. A new model was trained using 70% of the data and 15% of the data was used for internal validation. The remaining 15% was used for testing and radiologist review.
| Model A (Unilateral knee pain) |
10-Fold Cross Validation | ||
|---|---|---|---|
| Training | Validation | Testing | |
| n = 1505 | 90% | NA | 10% |
| Model B (Unilateral knee pain, WOMAC pain difference ≥ 3) |
10-Fold Cross Validation | ||
| Training | Validation | Testing | |
| n = 710 | 90% | NA | 10% |
| For radiologist review | Stratified Sampling | ||
| Training | Validation | Testing (for review) | |
| n = 710 | 70% (496) | 15% (107) | 15% (107) |
| Percent female | 59.2 | 62.0 | 60.1 |
| Age (years) | 61.0±9.3 | 60.5±9.0 | 6.06±9.1 |
| BMI (kg/m2) | 29.1±4.7 | 29.4±5.0 | 29.2±5.0 |
Model performance and statistical analysis
We performed 10-fold cross-validation on both models (Model A & B), and computed the area under curve (AUC) of the receiver operating characteristic (ROC) curves (Table 2). Descriptive statistics are presented as the mean along with the standard deviation. Unpaired Student’s t-test was used to compare the mean value of two different groups, and Fisher’s exact test was used to examine the non-random association between two groups of categorical variables. A p-value < 0.01 was considered statistically significant.
RESULTS
Among the 1,505 subjects from the baseline OAI (Model A in Table 1), the mean age was 60.7±9.1 years and the mean BMI was 28.7±4.7 kg/m2. About 56.9% of the selected subjects were women. When cases with WOMAC score difference <3 were excluded (Model B in Table 1), the sub-group demographic characteristics remained relatively similar with respect to the overall group (Mean age: 60.9±9.2 years; Mean BMI: 29.2±4.8 kg/m2 and Percentage women: 60.1). For cases that were reviewed by the radiologist (Table 1), stratified sampling based on age, BMI and gender helped us to generate a sub-group with demographic characteristics that were similar to the ones considered for models A & B.
The CAMs generated by extracting the features learned from the final convolutional layer of the neural network allowed us to examine the regions that were highly associated with knee pain. Previously, researchers who proposed CAMs used model architectures that had an in-plane resolution of 14×14 pixels (Figure 3A) [40]. We generated CAMs with higher resolution containing 28×28 pixels, which resulted in qualitatively improved identification of the regions associated with pain (Figure 3B). For comparison, we performed binary thresholding of the CAM-segmented areas for both cases. When a threshold of 0.5 was used on the CAMs, we found that only 6.0±2.4% of the overall image area was segmented from the CAMs generated by our model. In comparison, 22.3±9.4% of the image area was segmented when the same threshold was used on the CAMs generated by the model that was previously published [40].
Figure 3: CAMs on selected subjects within the test data.
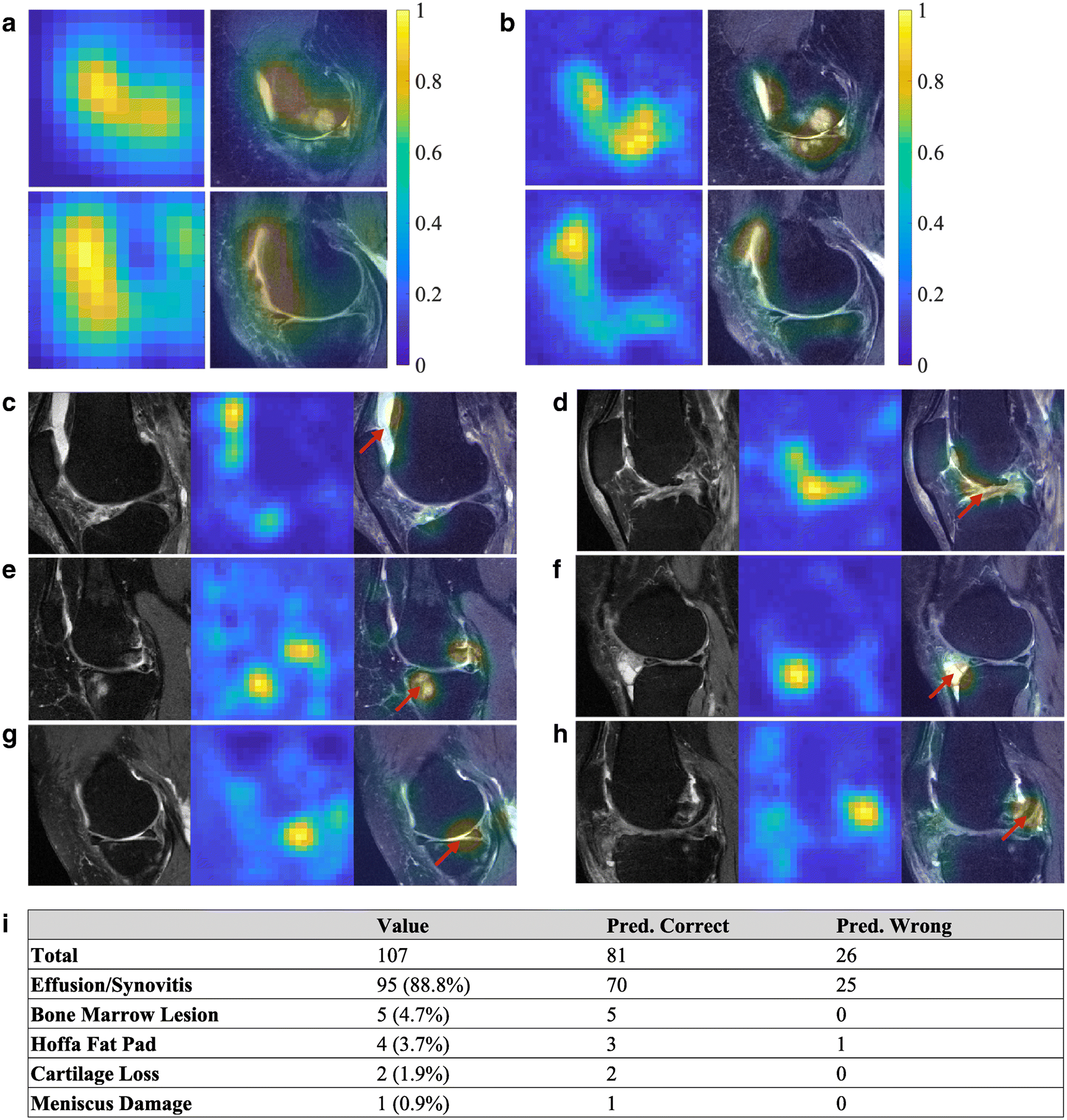
(A) Examples of CAMs generated from the fine-tuned VGGNet model, resulting in CAMs with an in-plane resolution of 14×14 pixels. (B) Examples of CAMs generated from the present model from the same MRI images with an in-plane resolution of 28×28 pixels. Both the heat maps and the overlap of the MR image with the heat map are shown. In some cases of the test data (C, D), effusion/synovitis was identified as the lesion present within the hot spots. Also, in few other cases, (E) bone marrow lesions, (F) Hoffa fat pad abnormality, (G) cartilage loss, and (H) meniscus damage, were identified as the lesions present within the hot spots. The red arrows indicate the locations of the identified structural regions. (I) Radiologist’s assessment on the test cases (n=107). For each case, the MR scan and the model-derived heat map of the knee with confirmed pain were reviewed by the radiologist, who then identified the presence of any lesions within the regions highlighted by the heat map.
After the radiologist review, the location and the type of lesions that were co-localized with the highlighted region in the CAMs of subjects were identified (Figures 3C–H). These lesions included effusion, synovitis, BML, Hoffa fat pad lesion, cartilage loss, and meniscal damage. The effusion on the selected intermediate-weighted MRI scans included effusion and synovitis, and therefore, we combined effusion/synovitis into a single category, as used in MRI Osteoarthritis Knee Score (MOAKS). Out of the 107 cases reviewed by the radiologist (Figure 3I), effusion/synovitis was found to be the most relevant structural abnormality related to frequent knee pain in 95 (88.8%) subjects. BML was found to be the most relevant abnormality for 5 (5.6%) subjects. Hoffa fat pad abnormalities were found for 4 (3.7%) subjects, cartilage loss was found for 2 (1.9%) subjects, and meniscal damage was found for 1 subject.
For the model trained with 1,505 subjects with unilateral knee pain (Model A), we observed an AUC of 0.808 on the test data (Figure 4A). For the case with 710 subjects with unilateral knee pain and with a difference in WOMAC pain score larger than 2 (Model B), the model achieved an AUC of 0.853 on the test data (Figure 4B). In comparison, models trained without image registration achieved AUCs of 0.769 and 0.812 on Model A and Model B, respectively.
Figure 4: Performance of the convolutional Siamese network model.
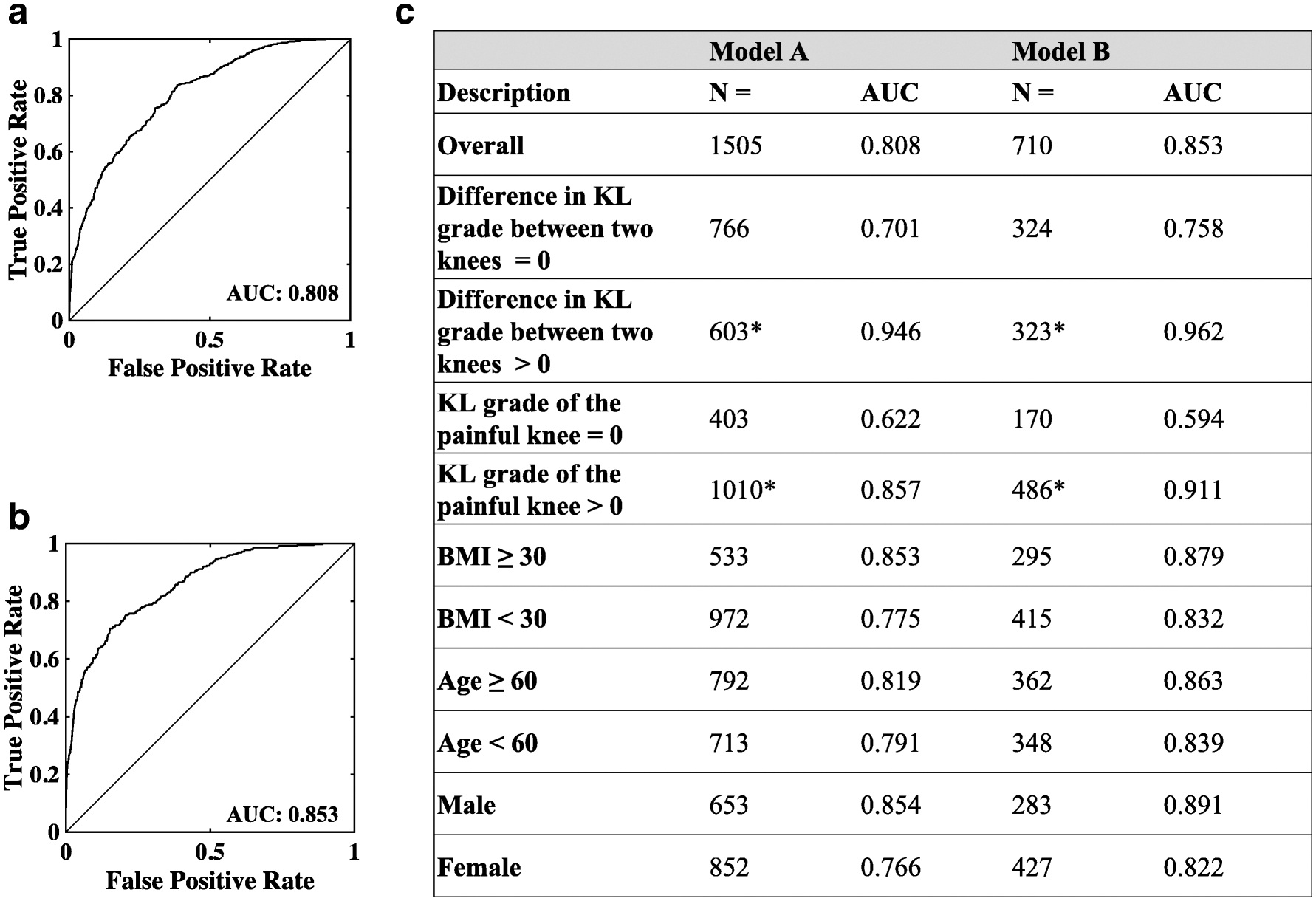
(A) Receiver operating characteristic (ROC) curve of Model A trained with 1,505 subjects with unilateral frequent knee pain. (B) ROC curve of Model B trained with 710 subjects with unilateral frequent knee pain and contralateral knee difference in WOMAC pain ≥ 3. (C) Sub-group analysis. Performance of the models as a function of KL-grade, BMI, age, and gender. The asterisk (*) is used to indicate that KL-grade was not available on few cases for the sub-group analysis.
Sub-group analysis further revealed that model performance varied with KL-grade, BMI, age and gender (Figure 4C). An interesting finding from our study is that our model could differentiate painful from nonpainful knees even when the OA disease stages were similar between the two knees (i.e. Difference in KL-grade = 0, AUC=0.701 for Model A and AUC=0.758 for Model B, Figure 4C). When there was a difference in radiographic OA between the knees (i.e. KL-grade difference ≠ 0), the model performance increased by about 26% for Model A and about 21% for Model B (Figure 4C). Our study also revealed that when the KL-grade of the painful knee was 0, the model resulted in a modest performance (AUC=0.622 for Model A and AUC=0.594 for Model B, Figure 4C). When the KL-grade was > 0, the model was able to better distinguish between the painful knee from the non-painful knee (Performance increased by 27% for Model A and about 35% for Model B, Figure 4C). It is also worthwhile to note that the WOMAC pain score of the painful knee was significantly lower for KL=0 cases than the KL>0 cases (3.10 vs 4.20, p≪0.01). The models also had higher values of AUC for subjects who were older, male and had higher BMI (Figure 4C). Specifically, we observed that subjects age 60 and older had a higher average KL-grade in the painful knee than the subjects who were younger than 60 years (1.71 vs 1.24; p<0.01). Also, males had higher averaged KL-grade in the painful knee than females (1.62 vs 1.37; p<0.01). Subjects with BMI greater than 30 also had higher averaged KL-grade in the painful knee than the subjects with lower BMI than 30 (1.8 vs 1.31; p≪0.01).
DISCUSSION
We developed a deep learning approach to distinguish painful knees from nonpainful contralateral ones, and we were able to do so with high accuracy. Further, an expert radiologist reviewing the knee areas that were identified as painful suggested that these were primarily regions of synovitis or effusion, which are known sources of knee pain in OA. Several studies have examined the association between radiographic features in the knee and knee pain within individual subjects and across multiple cohorts [15; 41–43]. While some studies relied on x-ray imaging, others relied on more sophisticated modalities such as MRI scans [44]. For some cases, associations between imaging features and unilateral pain were observed by comparing the knee with pain with the contralateral knee without pain in the same individual [45].
The Siamese network using both knees for the same individual to study unilateral pain effectively allowed one knee to serve as a control to the other knee with pain. Essentially, this dual-knee paradigm to assess knee pain is an attractive choice because the effect of common confounding factors such as age, gender and BMI, on the outcome of interest no longer applies in such scenarios. Also, unlike manual extraction of image-based or radiographic features which were then associated with knee pain, we investigated the feasibility of using deep learning to correlate structural regions from MRI scans of both knees with unilateral knee pain.
By combining information from a series of 2D slices, our model synthesized needed information from multiple locations to predict knee pain. This strategy resulted in a model that achieved high AUC (0.808), as evaluated using 10-fold cross validation. Model performance improved by about 5.6% when subjects with similar WOMAC pain score between the two knees were excluded (AUC=0.853). This improvement suggests that the Siamese network can identify the image features that are associated with a strong pain signature arising from one knee. This also implies that a set of image features that are common between the knees was also learned, but were not considered by the model to play a role in predicting knee pain. When compared to a machine learning approach using postero-anterior and lateral knee x-rays to predict knee pain, our model generated a significantly higher AUC in predicting unilateral knee pain [46]. We do however acknowledge that the training and testing datasets used for this study were different than the dataset presently used, and therefore a head-to-head comparison between the results is not feasible.
Our model’s improved performance when the difference in KL-grade was non-zero (Figure 4C), underscores the notion that prevalence and severity of knee pain is greatly influenced by the presence of pre-radiographic OA (KL=1) or radiographic OA (KL>1). This may imply that pain symptoms and potential structural abnormalities are perhaps less severe than the knees with frequent knee pain with pre-radiographic or radiographic OA. Indeed, previous studies have reported that knees of KL-grade 4 were 73–151 times more likely to have pain than knees of KL-grade 0 [44].
For the present study, we trained two identical neural networks in the ‘Siamese’ sense to generate a model that led to prediction of knee pain with high accuracy. In principle, pre-trained neural networks (i.e. VGGNet, AlexNet, etc.) can be used as part of the Siamese architecture. To evaluate this aspect, we generated another Siamese architecture using the network proposed by Zhou et. al. [40], for pairwise learning with the data split that was used for the radiologist review (Table 2). Interestingly, the AUC value on the test data for this case was 0.856. We also trained another Siamese architecture using a modified version of the AlexNet [40], as the neural network for pairwise learning. For this case, the AUC value on the test data was 0.863. In comparison, our proposed de novo Siamese network resulted in an AUC value of 0.862 on the test data. Results from these models imply that our proposed Siamese network achieved performance values that were similar to Siamese architectures based on pre-trained networks. However, the major difference is that our proposed network had high spatial resolution in the CAMs that allowed us to associate the results of CAMs with specific anatomical regions, and consequently to identify lesions that were highly correlated with knee pain.
While our deep learning model demonstrates promising results for predicting knee pain using MRI scans, there is room for improvement in model performance. Other network architectures can be explored such as deep autoencoders [47]. In a recent examination of x-rays and their prediction of knee pain, limiting the pain outcome to subjects who repeatedly reported knee pain increased the accuracy of x-ray prediction [46]. Alternate definitions of pain or tenderness could facilitate development of models with higher performance. Importantly, although postprocessing model results revealed effusion/synovitis to be the most common abnormality present within painful knees, we are by no means suggesting a causal pain mechanism.
In conclusion, this work demonstrates the use of a convolutional Siamese network that simultaneously associated MRI scans of an individual’s knees with unilateral knee pain. This framework allowed us to combine multiple 2D MRI slices from both the knees to efficiently construct the deep learning model. Our results provide a means by which to evaluate early imaging markers of OA and other joint disorders. Further validation of the deep learning model across different imaging datasets is necessary to validate this technique across the full spectrum of OA.
KEY POINTS.
Our article is the first to leverage a deep learning framework to associate MR images of the knee with knee pain.
We developed a convolutional Siamese network that had the ability to fuse information from multiple two-dimensional (2D) MRI slices from the knee with pain and the contralateral knee of the same individual without pain to predict unilateral knee pain.
Our model achieved an area under curve (AUC) value of 0.808. When individuals who had WOMAC pain scores that were not discordant for knees (pain discordance <3) were excluded, model performance increased to 0.853.
Funding Information:
This work was supported in part by the National Center for Advancing Translational Sciences, National Institutes of Health, through BU-CTSI Grant (1UL1TR001430), a Scientist Development Grant (17SDG33670323) from the American Heart Association, and a Research Award from the Hariri Institute for Computing and Computational Science & Engineering and Digital Health Initiative at Boston University, and NIH grants to VBK, DTF, and TDC (5U01AG-018820 & 1R01AR070139 and supported by the NIHR Manchester Biomedical Research Centre).
LIST OF ABBREVIATIONS
- AUC
Area Under Curve
- BMI
Body Mass Index
- BML
Bone Marrow Lesion
- CAM
Class Activation Mapping
- CNN
Convolutional Neural Network
- GPU
Graphical Processing Unit
- KL
Kellgren-Lawrence
- ML
Machine Learning
- MOAKS
MRI Osteoarthritis Knee Score
- MRI
Magnetic Resonance Imaging
- OA
Osteoarthritis
- OAI
Osteoarthritis Initiative
- ROC
Receiver Operating Characteristic
- WOMAC
Western Ontario and McMaster Universities Osteoarthritis Index
Footnotes
Publisher's Disclaimer: This Author Accepted Manuscript is a PDF file of a an unedited peer-reviewed manuscript that has been accepted for publication but has not been copyedited or corrected. The official version of record that is published in the journal is kept up to date and so may therefore differ from this version.
Guarantor:
The scientific guarantor of this publication is Dr. David Felson (dfelson@bu.edu).
Conflict of Interest:
Ali Guermazi is shareholder of BICL and consultant to Pfizer, AstraZeneca, TissueGene, Roche, Galapagos and MerckSerono.
Statistics and Biometry:
No complex statistical methods were necessary for this paper. Additionally, Dr. Kolachalama has needed statistical expertise.
Informed Consent:
Written informed consent was not required for this study because de-identified data is publicly available (Osteoarthritis Initiative: https://oai.nih.gov). At the time of enrollment, informed consent and ethical committee approvals were obtained by the OAI investigators.
Ethical Approval:
Institutional Review Board approval was not required because the data was obtained from a public database (Osteoarthritis Initiative: https://oai.nih.gov).
Study subjects or cohorts overlap:
The cohort has been reported in https://oai.nih.gov.
Methodology:
This is a case-control study.
REFERENCES
- 1.Cross M, Smith E, Hoy D et al. (2014) The global burden of hip and knee osteoarthritis: estimates from the global burden of disease 2010 study. Ann Rheum Dis 73:1323–1330 [DOI] [PubMed] [Google Scholar]
- 2.Neogi T (2013) The epidemiology and impact of pain in osteoarthritis. Osteoarthritis Cartilage 21:1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter DJ, Guermazi A, Roemer F, Zhang Y, Neogi T (2013) Structural correlates of pain in joints with osteoarthritis. Osteoarthritis Cartilage 21:1170–1178 [DOI] [PubMed] [Google Scholar]
- 4.Felson DT, Chaisson CE, Hill CL et al. (2001) The association of bone marrow lesions with pain in knee osteoarthritis. Ann Intern Med 134:541–549 [DOI] [PubMed] [Google Scholar]
- 5.Felson DT, Niu J, Guermazi A et al. (2007) Correlation of the development of knee pain with enlarging bone marrow lesions on magnetic resonance imaging. Arthritis Rheum 56:2986–2992 [DOI] [PubMed] [Google Scholar]
- 6.Hill CL, Gale DG, Chaisson CE et al. (2001) Knee effusions, popliteal cysts, and synovial thickening: association with knee pain in osteoarthritis. J Rheumatol 28:1330–1337 [PubMed] [Google Scholar]
- 7.Hill CL, Hunter DJ, Niu J et al. (2007) Synovitis detected on magnetic resonance imaging and its relation to pain and cartilage loss in knee osteoarthritis. Ann Rheum Dis 66:1599–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dimitroulas T, Duarte RV, Behura A, Kitas GD, Raphael JH (2014) Neuropathic pain in osteoarthritis: a review of pathophysiological mechanisms and implications for treatment. Semin Arthritis Rheum 44:145–154 [DOI] [PubMed] [Google Scholar]
- 9.Finan PH, Buenaver LF, Bounds SC et al. (2013) Discordance between pain and radiographic severity in knee osteoarthritis: findings from quantitative sensory testing of central sensitization. Arthritis Rheum 65:363–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clauw DJ, Witter J (2009) Pain and rheumatology: thinking outside the joint. Arthritis Rheum 60:321–324 [DOI] [PubMed] [Google Scholar]
- 11.O’Neill TW, Felson DT (2018) Mechanisms of Osteoarthritis (OA) Pain. Curr Osteoporos Rep 16:611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bedson J, Croft PR (2008) The discordance between clinical and radiographic knee osteoarthritis: a systematic search and summary of the literature. BMC Musculoskelet Disord 9:116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guermazi A, Zaim S, Taouli B, Miaux Y, Peterfy CG, Genant HG (2003) MR findings in knee osteoarthritis. Eur Radiol 13:1370–1386 [DOI] [PubMed] [Google Scholar]
- 14.Sowers MF, Hayes C, Jamadar D et al. (2003) Magnetic resonance-detected subchondral bone marrow and cartilage defect characteristics associated with pain and X-ray-defined knee osteoarthritis. Osteoarthritis Cartilage 11:387–393 [DOI] [PubMed] [Google Scholar]
- 15.Yusuf E, Kortekaas MC, Watt I, Huizinga TW, Kloppenburg M (2011) Do knee abnormalities visualised on MRI explain knee pain in knee osteoarthritis? A systematic review. Ann Rheum Dis 70:60–67 [DOI] [PubMed] [Google Scholar]
- 16.Wenham CY, Conaghan PG (2009) Imaging the painful osteoarthritic knee joint: what have we learned? Nat Clin Pract Rheumatol 5:149–158 [DOI] [PubMed] [Google Scholar]
- 17.Litjens G, Kooi T, Bejnordi BE et al. (2017) A survey on deep learning in medical image analysis. Med Image Anal 42:60–88 [DOI] [PubMed] [Google Scholar]
- 18.LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444 [DOI] [PubMed] [Google Scholar]
- 19.Hamm CA, Wang CJ, Savic LJ et al. (2019) Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 29:3338–3347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiryu S, Yasaka K, Akai H et al. (2019) Deep learning to differentiate parkinsonian disorders separately using single midsagittal MR imaging: a proof of concept study. Eur Radiol 29:6891–6899 [DOI] [PubMed] [Google Scholar]
- 21.Laukamp KR, Thiele F, Shakirin G et al. (2019) Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol 29:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vreemann S, Dalmis MU, Bult P et al. (2019) Amount of fibroglandular tissue FGT and background parenchymal enhancement BPE in relation to breast cancer risk and false positives in a breast MRI screening program : A retrospective cohort study. Eur Radiol 29:4678–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang CJ, Hamm CA, Savic LJ et al. (2019) Deep learning for liver tumor diagnosis part II: convolutional neural network interpretation using radiologic imaging features. Eur Radiol 29:3348–3357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norman B, Pedoia V, Majumdar S (2018) Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology 288:177–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ambellan F, Tack A, Ehlke M, Zachow S (2019) Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: Data from the Osteoarthritis Initiative. Med Image Anal 52:109–118 [DOI] [PubMed] [Google Scholar]
- 26.Gaj S, Yang M, Nakamura K, Li X (2019) Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks. Magn Reson Med. 10.1002/mrm.28111 [DOI] [PubMed] [Google Scholar]
- 27.Byra M, Wu M, Zhang X et al. (2020) Knee menisci segmentation and relaxometry of 3D ultrashort echo time cones MR imaging using attention U-Net with transfer learning. Magn Reson Med 83:1109–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tack A, Mukhopadhyay A, Zachow S (2018) Knee menisci segmentation using convolutional neural networks: data from the Osteoarthritis Initiative. Osteoarthritis Cartilage 26:680–688 [DOI] [PubMed] [Google Scholar]
- 29.Prasoon A, Petersen K, Igel C, Lauze F, Dam E, Nielsen M (2013) Deep feature learning for knee cartilage segmentation using a triplanar convolutional neural network. Med Image Comput Comput Assist Interv 16:246–253 [DOI] [PubMed] [Google Scholar]
- 30.Bien N, Rajpurkar P, Ball RL et al. (2018) Deep-learning-assisted diagnosis for knee magnetic resonance imaging: Development and retrospective validation of MRNet. PLoS Med 15:e1002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Englund M, Niu J, Guermazi A et al. (2007) Effect of meniscal damage on the development of frequent knee pain, aching, or stiffness. Arthritis & Rheumatism 56:4048–4054 [DOI] [PubMed] [Google Scholar]
- 32.Felson DT, Niu J, Guermazi A et al. (2007) Correlation of the development of knee pain with enlarging bone marrow lesions on magnetic resonance imaging. Arthritis & Rheumatism 56:2986–2992 [DOI] [PubMed] [Google Scholar]
- 33.Hill CL, Hunter DJ, Niu J et al. (2007) Synovitis detected on magnetic resonance imaging and its relation to pain and cartilage loss in knee osteoarthritis. Annals of the Rheumatic Diseases 66:1599–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckstein F, Benichou O, Wirth W et al. (2009) Magnetic resonance imaging-based cartilage loss in painful contralateral knees with and without radiographic joint space narrowing: Data from the osteoarthritis initiative. Arthritis & Rheumatism 61:1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cibere J, Sayre EC, Guermazi A et al. (2011) Natural history of cartilage damage and osteoarthritis progression on magnetic resonance imaging in a population-based cohort with knee pain. YJOCA 19:683–688 [DOI] [PubMed] [Google Scholar]
- 36.Kim HA, Kim I, Song YW et al. (2011) The association between meniscal and cruciate ligament damage and knee pain in community residents. YJOCA 19:1422–1428 [DOI] [PubMed] [Google Scholar]
- 37.Peterfy CG, Schneider E, Nevitt M (2008) The osteoarthritis initiative: report on the design rationale for the magnetic resonance imaging protocol for the knee. Osteoarthritis Cartilage 16:1433–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fawaz-Estrup F (2004) The osteoarthritis initiative: an overview. Med Health R I 87:169–171 [PubMed] [Google Scholar]
- 39.Chopra S, Hadsell R, 1 YLC, 2005. Learning a similarity metric discriminatively, with application to face verification. csutorontoca [Google Scholar]
- 40.Zhou B, Khosla A, Lapedriza A, Oliva A, Torralba A (2016) Learning Deep Features for Discriminative Localization. 2016 Ieee Conference on Computer Vision and Pattern Recognition (Cvpr). 10.1109/Cvpr.2016.319:2921-2929 [DOI] [Google Scholar]
- 41.Bedson J, Croft PR (2008) The discordance between clinical and radiographic knee osteoarthritis: A systematic search and summary of the literature. BMC Musculoskeletal Disorders 9:467–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wenham CY, Conaghan PG (2009) Imaging the painful osteoarthritic knee joint: what have we learned? Nature Reviews Rheumatology 5:149–158 [DOI] [PubMed] [Google Scholar]
- 43.Sayre EC, Guermazi A, Esdaile JM et al. (2017) Associations between MRI features versus knee pain severity and progression: Data from the Vancouver Longitudinal Study of Early Knee Osteoarthritis. PLOS ONE 12:e0176833–0176812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Neogi T, Felson D, Niu J et al. (2009) Association between radiographic features of knee osteoarthritis and pain: results from two cohort studies. BMJ 339:b2844–b2844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Javaid MK, Kiran A, Guermazi A et al. (2012) Individual magnetic resonance imaging and radiographic features of knee osteoarthritis in subjects with unilateral knee pain: The Health, Aging, and Body Composition Study. Arthritis & Rheumatism 64:3246–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minciullo L, Parkes MJ, Felson DT, Cootes TF (2018) Comparing image analysis approaches versus expert readers: the relation of knee radiograph features to knee pain. Ann Rheum Dis 77:1606–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dolz J, Desrosiers C, Ben Ayed I (2018) 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. NeuroImage 170:456–470 [DOI] [PubMed] [Google Scholar]


