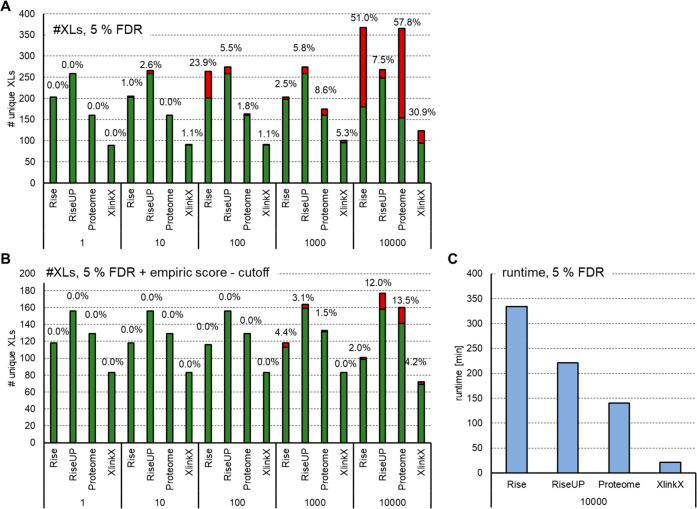
Figure 4.
Comparison of MeroX in different modes and XlinkX upon the variation of database size. (A) DSBU cross-linked BSA was analyzed using MeroX (v 2.0.1.4, different modes) or XlinkX (within Thermo Proteome discoverer v 2.4) against a database containing BSA spiked with proteins of the human proteome to obtain total sizes of 1–10 000 proteins. Results were filtered at 5% FDR on the spectrum level. Bars indicate the number of unique cross-linked residue pairs: green, BSA (intra-) links; red, interlinks and non-BSA intralinks. (B) In addition, a score cutoff of 50 was applied to all MeroX results, and XlinkX data were filtered for a minimal score of 40 and a minimal delta score of 4. (C) Runtime needed for data analysis with the largest database containing 10 000 proteins.