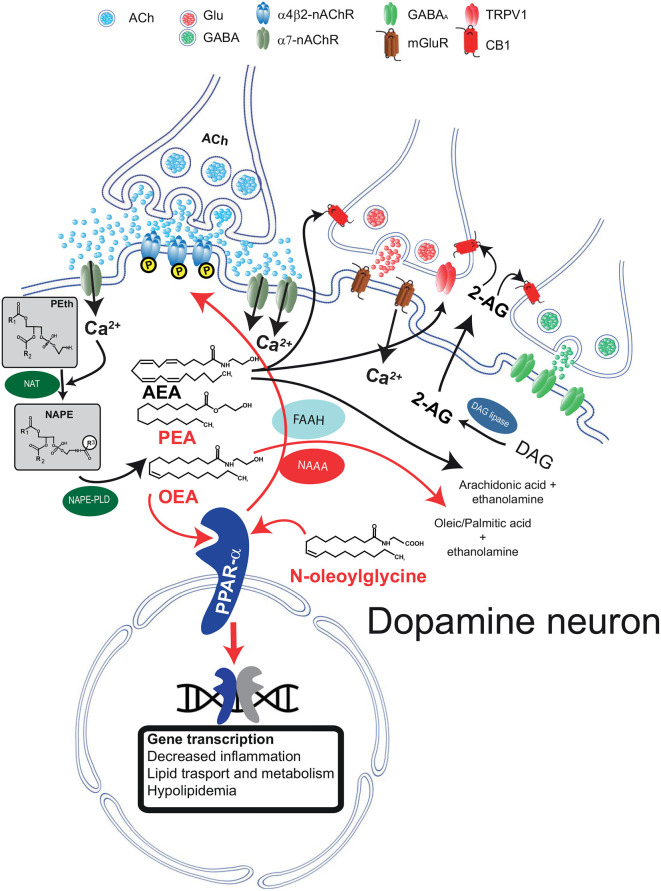
Figure 1.
Schematic diagram illustrating the biosynthetic and catabolic pathways for N-acylethanolamine (NAE) and canonical endocannabinoid formation and catabolism, and their cellular mechanisms of actions through their receptors. Phosphatidylethanolamine (Peth) is converted into N-acyl-phosphatidylethanolamine (NAPE) by N-acyltransferase (NAT). Ca2+ entry mediated by α7-nAChRs activates NAEs synthesis through the Ca2+ dependent NAT. The resulting NAPE is hydrolyzed by NAPE-PLD to the corresponding NAEs anandamide (AEA), oleoyl ethanolamide (OEA), and palmitoylethanolamide (PEA). Activation of PPARα by NAEs results in genomic effects (gene transcription) and in non-genomic actions, such as activation of a tyrosine kinase and phosphorylation of β2*nAChRs (i.e., α4β2). Fatty acid amide hydrolase (FAAH) and NAE-hydrolyzing acid amidase (NAAA) are the major inactivating enzymes for OEA, PEA, and AEA and convert them in ethanolamine and corresponding fatty acids (oleic, palmitic, and arachidonic acids, respectively). NAAA preferentially hydrolyzes PEA. N-oleoyl glycine is one member of the N-acyl amino acid family and is known to activate PPARα. The figure illustrates that AEA and 2-arachidonoylglycerol (2-AG) are produced on demand by NAPE-PLD and DAG lipase, respectively. Raises in intracellular Ca2+ can be induced, as in the example, by activation of metabotropic glutamate receptors (mGluR). 2-AG and AEA bind to presynaptic CB1 receptors expressed on GABA and glutamate terminals and depress neurotransmitter release. AEA also activates TRPV1 receptors located on presynaptic glutamatergic terminals. Abbreviations: NAPE-PLD, N-acyl phosphatidylethanolamine phospholipase D; DAG, diacylglycerol; MAG, monoacylglycerol; FAAH, fatty acid amide hydrolase; Glu, glutamate; CB1, cannabinoid type-1 receptor; TRPV1, transient receptor potential vanilloid type-1; PPARα, peroxisome proliferator-activated receptor type-α; nAChRs, nicotinic acetylcholine receptors. This figure is adapted, with permission, from Melis and Pistis (2012) and Pistis and Muntoni (2017).