This retrospective cohort study reveals discordant findings to adult pharmacogenetic guidelines for sertraline, escitalopram, and citalopram and CYP2C19 necessitating further investigation before application to pediatrics.
Abstract
Video Abstract
OBJECTIVES:
To determine the association between cytochrome P450 2C19 (CYP2C19) metabolizer status and risk for escitalopram and citalopram, collectively termed (es)citalopram, and sertraline adverse events (AEs) in children.
METHODS:
In this retrospective cohort study, we used deidentified electronic health records linked to DNA. The cohort included children ≤18 years with ≥2 days of (es)citalopram or ≥7 days of sertraline exposure. The primary outcome was AEs assessed by manual chart review. CYP2C19 was genotyped for functional variants (*2, *3, *4, *6, *8, and *17), and individuals were assigned metabolizer status. Association between AEs and metabolizer status was determined by using Cox regression adjusting for age, race, ethnicity, dose, and concomitant CYP2C19-inhibiting medications.
RESULTS:
The cohort included 249 sertraline-exposed and 458 (es)citalopram-exposed children, with a median age of 14.2 years (interquartile range 11.2–16.2) and 13.4 years (interquartile range 10.1–15.9), respectively. Sertraline AEs were more common in normal metabolizers (NMs) compared to poor metabolizers (PMs) or intermediate metabolizers (IMs) (hazard ratio [HR] 1.8; 95% confidence interval [CI] 1.01–3.2; P = .047) in unadjusted analysis and after adjustment (HR 1.9; CI 1.04–3.4; P = .04). For (es)citalopram, more AEs were observed in NMs than PMs and IMs without statistically significant differences (unadjusted HR 1.6; CI 0.95-2.6; P = .08; adjusted HR 1.6; CI 0.95-2.6; P = .08).
CONCLUSIONS:
In contrast to adults, in our pediatric cohort, CYP2C19 NMs experienced increased sertraline AEs than PMs and IMs. (Es)citalopram AEs were not associated with CYP2C19 status in the primary analysis. The mechanism underlying this pediatric-specific finding is unknown but may be related to physiologic differences of adolescence. Further research is required to inform genotype-guided prescribing for these drugs in children.
What’s Known on This Subject:
Adult studies suggest that CYP2C19 poor metabolizers have increased risk for sertraline, citalopram, and escitalopram adverse events, although previous pediatric studies do not consistently support this association.
What This Study Adds:
This is the first pediatric study to suggest that in contrast to adults, children with decreased or no CYP2C19 activity are at decreased risk for sertraline adverse events compared with those with normal metabolism.
Selective serotonin reuptake inhibitors (SSRIs) such as escitalopram and citalopram, collectively termed (es)citalopram, and sertraline increase serotonergic activity by decreasing presynaptic serotonin reuptake.1 SSRIs are the most common antidepressant class used in pediatric patients for a variety of indications.2 Adverse events (AEs) to SSRIs have been reported to be more frequent in younger children than in adults, and authors of previous studies have reported that 5% to 32% of SSRI-exposed children experience an AE.3 Children experience a distinct spectrum of AEs compared with adults and are more likely to have activation.4,5
Sertraline and (es)citalopram are primarily metabolized by the cytochrome P450 2C19 (CYP2C19) enzyme, whereas other enzymes such as cytochrome P450 2D6 contribute to a lesser extent.1 Interindividual differences in CYP2C19 function can be due to common genetic variants in CYP2C19 that lead to a spectrum of activity, including no function in poor metabolizers (PMs) to increased function in ultrarapid metabolizers (UMs).6 The Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines recommend considering a 50% dose reduction for CYP2C19 PMs after initiation of sertraline or (es)citalopram.6 However, given limitations to what is known about CYP2C19 function and this drug-gene interaction in children, it is recommended to use caution when extrapolating these guidelines to the pediatric population.6 Considering the paucity of pharmacogenetic data for sertraline and (es)citalopram in children and adolescents, we performed a retrospective observational study using biobanked DNA linked to deidentified electronic health record (EHR) data. Our hypothesis was that in a pediatric cohort treated with the SSRIs of interest during 1998–2019 with variable follow-up, those with poor or intermediate CYP2C19 enzyme activity have increased incidence of AEs compared with individuals who have normal CYP2C19 enzyme function.
Methods
Study Design
This was a retrospective cohort study with variable follow-up. Data for this study were collected from BioVU, the Vanderbilt University Medical Center (VUMC) biobank that links DNA to deidentified EHRs.7–9 This study was reviewed by the Vanderbilt Institutional Review Board and determined to be non–human subjects research. The study cohorts were identified through an initial automated search, followed by manual review to confirm further inclusion and exclusion criteria. The automated search criteria included age <18 years at the time of first sertraline or (es)citalopram mention between 1998 and 2019, at least 2 mentions of sertraline at a minimum of 7 days apart or at least 2 mentions of (es)citalopram at a minimum of 2 days apart, and a noncompromised DNA sample in BioVU. Additional inclusion criteria assessed by manual review were at least 1 note from a VUMC provider managing the SSRI on or after day 7 of a new sertraline course and on or after day 2 of (es)citalopram therapy to ensure documentation of AEs. Exclusion criteria were lack of notes indicating medical decision-making or management of the SSRI and lack of documentation regarding the presence or absence of AEs.
Individuals who met inclusion criteria entered the cohort on day 1 of the new sertraline or (es)citalopram prescription. Individuals permanently exited the cohort by being censored at the earliest occurrence of the following: AE, discontinuation of the medication, death, no medication management notes available for 1 year, end of follow-up period occurring, or if the individual completed 500 days of therapy. Time in the cohort was measured in days.
Outcome Measures and Identification
Outcomes were determined blinded to CYP2C19 genotype and phenotype. The primary outcome was AEs in subjects taking sertraline or (es)citalopram. AEs were defined as any untoward event reported by the patient, caregiver, or provider suspected or confirmed to be due to sertraline or (es)citalopram when the medication was taken in the prescribed and intended manner that resulted in a clinically significant action including 1 of the following: unexpected medical care, decreased SSRI dosage, or cessation of the medication. If AEs were noted, details regarding the AE were noted, including the type of AE, date of AE, dose of SSRI at the AE, and management by the prescriber. AEs were identified through manual review of each individual’s EHR with a separate reviewer for sertraline and (es)citalopram. An additional reviewer independently reviewed 25 records from the sertraline and (es)citalopram cohorts to determine AE presence or absence, blinded to metabolizer status and the other reviewer’s determination to calculate an interrater reliability of AE status.
Data Abstraction
Data from this study were collected and stored in Research Electronic Data Capture, an electronic management tool hosted by VUMC.10 The following data were extracted manually by a single reviewer for each individual in the study cohort: demographic data (sex, race, ethnicity, age, and weight at initial dose of SSRI), pertinent clinical information (indication for SSRI and mental health diagnoses), medication data (SSRI dosage amount, SSRI duration, and concomitant drugs), and presence or absence of AEs. Concomitant medications were defined as prescription medications and regular use of over-the-counter medications on the day of initiation as noted in the closest note and/or from medication administration data. Use of concomitant CYP2C19-inhibiting medications by strength (strong, moderate, weak, or to be determined [TBD]) and CYP2C19-inducing medications were noted as defined from the Flockhart table.11
DNA Analysis
CYP2C19 genotyping was performed on each individual’s DNA by the Vanderbilt Technologies for Advanced Genomics laboratory by using reagents and protocols as specified by the manufacturer. Genotyping was performed by using 6 TaqMan assays (Life Technology, Forest City, CA) to determine the presence or absence of CYP2C19 no-function alleles (*2 [rs4244285], *3 [rs4986893], *4 [rs28399504], *6 [rs72552267], and *8 [rs41291556]) or increased-function alleles (*17, rs12248560). A normal-function allele (*1) was assigned if none of the tested alleles were identified. CYP2C19 metabolizer status was assigned (PM, intermediate metabolizer [IM], normal metabolizer [NM], rapid metabolizer [RM], or UM) according to CPIC guidelines (Supplemental Table 3).12,13
Statistical Analysis
Primary analysis was performed by comparing CYP2C19 PMs and IMs to NMs. For this analysis, PMs were combined with IMs because of the small number of PMs. All demographic, clinical, and medication outcomes were calculated as frequencies and percentages for categorical variables or medians and interquartile ranges (IQRs) for continuous variables. For the primary analysis, comparisons of AEs between PMs and IMs versus NMs were performed by using Cox regression to account for variable follow-up for individuals taking sertraline and (es)citalopram. A multivariate Cox regression was performed for individuals taking sertraline and (es)citalopram among PMs/IMs and NMs with AEs as the primary outcome, adjusting for age, race, ethnicity, dose, and concomitant use of any type of CYP2C19-inhibiting medications (including strong, moderate, weak, and TBD strengths). Secondary analysis was performed comparing AEs by using 3 metabolizer groups (PMs and IMs, NMs, and RMs and UMs) for individuals on sertraline and (es)citalopram by using Cox regression in univariate and multivariate analysis adjusting for the same covariates. Exploratory analyses are described in Supplemental Information and Supplemental Tables 3–6. Data analysis was performed by using Stata version 15.1 (Stata Corp, College Station, TX). Any P value <.05 was considered to be statistically significant, and all statistical tests were 2 sided.
Results
Study Cohorts and CYP2C19 Analysis
Sertraline
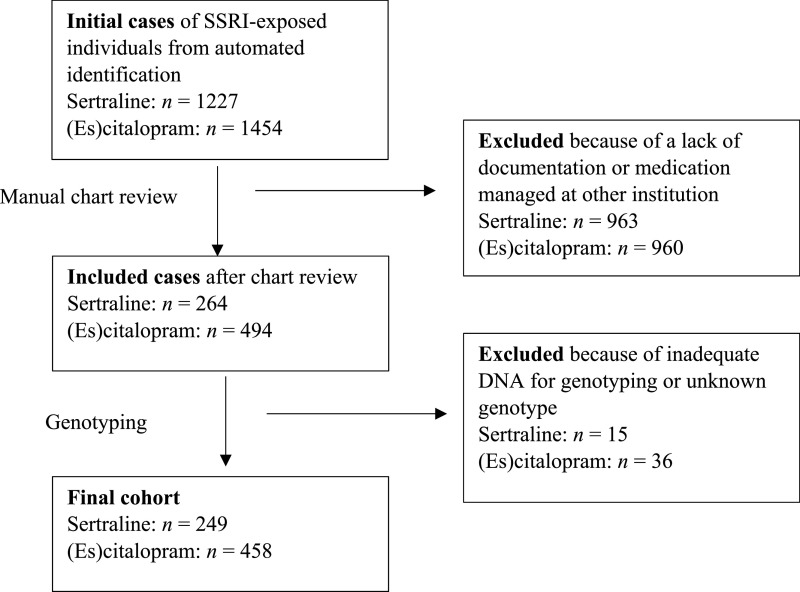
The automated search identified 1227 sertraline-exposed individuals, of whom 249 were included in the final cohort (Fig 1, Supplemental Table 4). The median age was 14 years (IQR 11–16), and most were female (154 [61.9%]), white (217 [87.2%]), and non-Hispanic (240 [96.4%]) (Table 1). In all, 12 (4.8%) were PMs, 72 (28.9%) were IMs, 88 (35.4%) were NMs, 66 (26.5%) were RMs, and 11 (4.4%) were UMs. CYP2C19 allele frequencies in our cohort were consistent with expected frequencies.6 A total of 74 individuals (29.7%) were taking any concomitant CYP2C19-inhibiting medication at the start of sertraline therapy (Table 1, Supplemental Table 5). Demographic variables, indications for sertraline, presence of comorbidities, dosing regimen, and concomitant use of any CYP2C19-inhibiting medications were similar across metabolizer status except for ethnicity (Table 2).
FIGURE 1.
Study overview.
TABLE 1.
Demographics, Metabolizer Status, and Sertraline Exposures in Study Cohort by Medication
| Sertraline (n = 249) | (Es)citalopram (n = 458) | |
|---|---|---|
| Age at SSRIa initiation, y, median (IQR) | 14.2 (11.2–16.2) | 13.4 (10.1–15.9) |
| Sex, n (%) | ||
| Male | 95 (38.1) | 196 (42.8) |
| Female | 154 (61.9) | 262 (57.2) |
| Race, n (%) | ||
| White | 217 (87.2) | 378 (82.5) |
| African American | 20 (8.0) | 41 (9.0) |
| Asian or Pacific Islander | 2 (0.8) | 7 (1.5) |
| Native American | 1 (0.4) | 0 |
| Unknown | 7 (2.8) | 30 (6.6) |
| Other | 2 (0.8) | 2 (0.4) |
| Ethnicity, n (%) | ||
| Hispanic | 9 (3.6) | 25 (5.5) |
| Non-Hispanic | 240 (96.4) | 433 (94.5) |
| Initial SSRIa dose, mg/d, median (IQR) | 25 (12.5–25) | 10 (5–10) |
| CYP2C19 metabolizer phenotype, n (%) | ||
| PM | 12 (4.8) | 8 (1.8) |
| IM | 72 (28.9) | 132 (28.8) |
| NM | 88 (35.4) | 183 (40.0) |
| RM | 66 (26.5) | 115 (25.1) |
| UM | 11 (4.4) | 20 (4.4) |
| Concomitant use of any CYP2C19-inhibiting medications, n (%) | 74 (29.7) | 104 (22.7) |
| AE, n (%) | 65 (26.1) | 111 (24.4) |
TABLE 2.
Comparison of Demographic and Baseline Characteristics Among Metabolizer Groups
| Sertraline | (Es)citalopram | |||||||
|---|---|---|---|---|---|---|---|---|
| CYP2C19 PMs/IMs (n = 84) | CYP2C19 NMs (n = 88) | Pa | CYP2C19 RMs/UMs (n = 77) | CYP2C19 PMs/IMs (n = 140) | CYP2C19 NMs (n = 183) | Pa | CYP2C19 RMs/UMs (n = 135) | |
| Age, y, median (IQR) | 14.1 (10.5–16.3) | 14.4 (12.1–16.5) | .36 | 13.8 (11.5–16) | 13.6 (9.7–15.9) | 12.9 (9.9–15.5) | .65 | 14.2 (10.3–16.1) |
| Female sex, frequency (%) | 55 (65.0) | 52 (59.1) | .43 | 47 (61.0) | 83 (59.3) | 102 (55.7) | .58 | 77 (57.0) |
| Race, frequency (%) | .14 | .65 | ||||||
| White | 72 (85.7) | 77 (87.5) | — | 68 (88.3) | 116 (82.9) | 148 (80.9) | — | 114 (84.4) |
| African American | 9 (10.7) | 3 (3.4) | — | 8 (10.4) | 10 (7.1) | 16 (8.7) | — | 15 (11.1) |
| Asian or Pacific Islander | 1 (1.2) | 1 (1.1) | — | 0 | 4 (2.9) | 3 (1.6) | — | 0 |
| Native American | 0 | 1 (1.1) | — | 0 | 0 | 0 | — | 0 |
| Unknown | 1 (1.2) | 5 (5.7) | — | 1 (1.3) | 9 (6.4) | 16 (8.7) | — | 5 (3.7) |
| Other | 1 (1.2) | 1 (1.1) | — | 0 | 1 (0.7) | 0 | — | 1 (0.7) |
| Ethnicity, frequency (%) | .04 | 0.99 | ||||||
| Hispanic | 1 (1.2) | 8 (9.9) | — | 0 | 9 (6.4) | 12 (6.6) | — | 4 (3.0) |
| Non-Hispanic | 83 (98.1) | 80 (90.9) | — | 77 (100.0) | 131 (93.6) | 171 (93.4) | — | 131 (97.0) |
| Indication, frequency (%) | ||||||||
| Anxiety | 33 (39.3) | 44 (50.0) | .17 | 38 (49.4) | 75 (53.6) | 99 (54.1) | 0.99 | 74 (54.8) |
| Depression | 34 (40.5) | 33 (37.5) | .76 | 27 (35.1) | 79 (56.4) | 83 (45.4) | .06 | 75 (55.6) |
| Psychiatric comorbidities, frequency (%) | 44 (52.5) | 50 (56.8) | 0.65 | 46 (59.7) | 39 (27.9) | 38 (20.8) | .15 | 27 (20.0) |
| Use of any CYP2C19-inhibiting medications, frequency (%) | 28 (33.3) | 28 (31.8) | .87 | 18 (23.4) | 29 (20.7) | 39 (21.3) | 0.99 | 36 (26.7) |
| Initial dose, mg/d, median (IQR) | 25 (12.5–25) | 25 (12.5–25) | .42 | 25 (12.5–25) | 10 (5–10) | 10 (5–10) | .77 | 10 (5–10) |
| AEs, frequency (%) | 18 (21.4) | 31 (35.2) | .047 | 16 (20.8) | 24 (17.1) | 46 (25.1) | .10 | 41 (30.4) |
—, not applicable.
P values for CYP2C19 PMs and IMs versus NMs from Kruskal-Wallis test for continuous variables and Fisher’s exact or Pearson’s χ2 for categorical variables.
(Es)citalopram
Of 1454 (es)citalopram-exposed individuals identified by the automated search, 458 were included in the final cohort (Fig 1, Supplemental Table 4). The median age was 13 years (IQR 10–16), and most were female (262 [57.2%]), white (378 [82.5%]), and non-Hispanic (433 [94.5%]) (Table 1). With respect to CYP2C19 metabolizer status 8 (1.8%) were PMs, 132 (28.8%) were IMs, 183 (40.0%) were NMs, 115 (25.1%) were RMs, and 20 (4.4%) were UMs, consistent with previously reported frequencies.6 A total of 104 (22.5%) were taking any concomitant CYP2C19-inhibiting medication at (es)citalopram start (Table 1, Supplemental Table 5). All data for demography, indication for (es)citalopram, comorbidities, dosing regimen, and concomitant CYP2C19-inhibiting medications were similar across metabolizer status (Table 2).
AEs
Sertraline
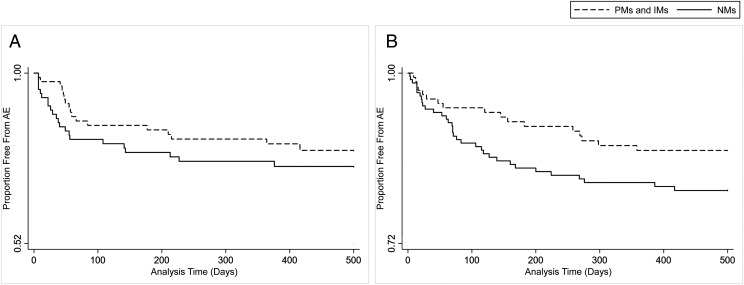
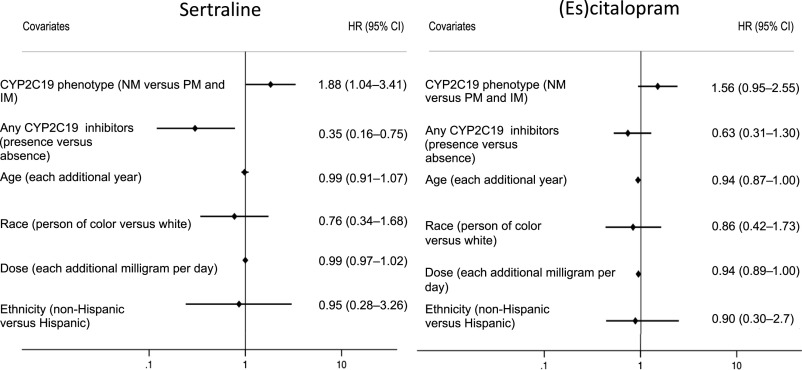
In total, 65 (26.1%) individuals exposed to sertraline experienced 88 AEs. The κ-statistic for interrater reliability of AE status, on the basis of independent blinded review, was 0.7, indicating substantial agreement. There were 31 different types of AEs, and the most common were activation, worsened mood or irritability, agitation, and somnolence. A complete list of AE types and frequencies are listed in Supplemental Table 7. The time to AE survival analysis reveals an earlier time to AE in NMs compared to PMs and IMs, as displayed in Fig 2. AE frequency across all metabolizer classes is shown in Supplemental Fig 4. In univariate analysis, the risk of sertraline AEs was higher in NMs compared to PMs and IMs (hazard ratio [HR] 1.8; 95% confidence interval [CI] 1.01–3.2; P = .047). In multivariable analysis, after adjusting for age, race, ethnicity, dose, and concomitant use of any CYP2C19-inhibiting medications, the risk for sertraline AEs remained higher in NMs compared to PMs and IMs (HR 1.9; 95% CI 1.04–3.4; P = .04; Fig 3). In this model, the presence of any CYP2C19-inhibiting medication was also significant (HR 0.4; 95% CI 0.2–0.8; P = .007) in the same direction as metabolizer status, with those not taking a CYP2C19 inhibitor having more AEs. Exploratory analysis comparing AEs among CYP2C19 phenotypic activity scores (incorporating genotype and concomitant medications) was not significant (Supplemental Information). Secondary analysis was performed to compare AEs across all metabolizer classes. There was no association of CYP2C19 metabolizer status and sertraline AEs when all 3 CYP2C19 metabolizer status groupings were included (ie, PMs and IMs, NMs, and RMs, UMs) in univariate (HR 1.0; 95% CI 0.7–1.3; P = .78) or multivariate analysis adjusting for age, dose, ethnicity, race, and concomitant use of CYP2C19-inhibiting medications (HR 0.9; 95% CI 0.7–1.3; P = .68). Exploratory analyses were notable for RMs and UMs having a lower risk for AEs than NMs in multivariate analysis (HR 0.5; 95% CI 0.2–0.9; P = .02) when adjusting for start dose, ethnicity, race, age, and any concomitant CYP2C19-inhibiting medications.
FIGURE 2.
AEs among CYP2C19 PMs and IMs (dashed line) and NMs (solid line) for sertraline (HR 1.8; 95% CI 1.01–3.2; P = .047) and (es)citalopram (HR 1.6; CI 0.95–2.6; P = .08). A, Sertraline time to AE. B, (Es)citalopram time to AE.
FIGURE 3.
Cox regression of AEs. For sertraline, NM status was associated with increased AE risk, and use of CYP2C19-inhibiting medications was associated with decreased risk. For (es)citalopram, younger age was associated with increased AE risk, and CYP2C19 phenotype and concomitant inhibitor use revealed no statistical significance.
(Es)citalopram
Among children taking (es)citalopram, 111 (24.4%) children experienced 198 AEs. The κ-statistic for interrater reliability of AE status was 0.9, indicating excellent agreement. The most common AEs were activation, worsened mood, aggression, and somnolence (Supplemental Table 7). Figure 2 and Supplemental Figure 4 reveal (es)citalopram time to AE by CYP2C19 metabolizer phenotype across PMs/IMs and NMs, as well as across each individual metabolizer class, respectively. In the primary analysis, there were more (es)citalopram AEs in NMs compared to PMs and IMs in univariate analysis, but this did not reach statistical significance (HR 1.6; 95% CI 0.95–2.6; P = .08). After adjustment for age, race, ethnicity, dose, and concomitant use of any CYP2C19-inhibiting medications, the association of (es)citalopram AEs in NMs compared to PMs and IMs was again not statistically significant (HR 1.6; 95% CI 0.95–2.6; P = .08; Fig 3). A secondary analysis was performed comparing all metabolizer classes and revealed an increased risk of (es)citalopram AEs in those with increased metabolism among PMs and IMs, NMs, and RMs, UMs (HR 1.4; 95% CI 1.1–1.8; P = .01) in univariate Cox regression. This association was also present in multivariate secondary analysis, as Cox regression adjusting for age, race, ethnicity, dose, and concomitant use of any CYP2C19-inhibiting medications revealed more (es)citalopram AEs in those with faster metabolism (HR 1.4; 95% CI 1.1–1.8; P = .006). The results of exploratory analyses for (es)citalopram are listed in the Supplemental Information.
Discussion
In this pediatric cohort, we expected to find more SSRI AEs among CYP2C19 PMs and IMs, consistent with the known metabolic pathways for these drugs and published adult data. Instead, our data reveal more AEs in children and adolescents treated with sertraline in CYP2C19 NMs compared to PMs and IMs in both univariate and multivariate analyses. In this cohort, the presence of any CYP2C19-inhibiting medication also resulted in a lower risk for AEs compared with children who were not taking an inhibiting medication. For (es)citalopram, we similarly did not see the expected increased AEs among PMs and IMs but rather observed no association of CYP2C19 metabolizer status to (es)citalopram AEs. These findings, discordant from adults, indicate that the drug-gene interactions for CYP2C19 and SSRIs may differ in children and adolescents versus adults.
Our data are discordant with previous adult and pediatric studies in which authors examine sertraline AEs and CYP2C19. There are guidelines for sertraline dosing that are based on CYP2C19 metabolizer status for adults, which suggest extrapolation to children, although pediatric data are limited.6,14 In 1 previous pediatric study in 352 participants, researchers found no association of CYP2C19 metabolizer status with the total number of sertraline side effects experienced.15 The AE definition in that study (namely, algorithmic identification of keywords for 10 common sertraline AEs) led to a high AE rate (95%) which may have precluded observing differences across groups.15 Our study required EHR documentation that (1) the SSRI was suspect to cause the AE and (2) a clinically significant action occurred (eg, decreased dose), leading to a lower AE rate (26%). In a study of sertraline efficacy in children with fragile X, authors reported higher levels of clinical functioning compared to the baseline in CYP2C19 PMs and IMs, but AEs were not assessed.16 With this study, we are the first to observe a unique association of CYP2C19 function and sertraline AEs in children.
Our study data are also discordant with previous adult and pediatric data on the association of AEs and (es)citalopram. One previous retrospective pediatric study with 263 children and adolescents on (es)citalopram with anxiety and/or depression revealed increased AEs in those with reduced or no CYP2C19 metabolism compared with those with normal or fast metabolism.17 This contrasts a smaller pharmacokinetic study with 19 children or adults, which revealed no effect of decreased CYP2C19 or CYP2D6 function on citalopram metabolites, although the number with reduced CYP2C19 metabolism was small (3 IMs).18 In sum, the evidence regarding the association of (es)citalopram AEs and CYP2C19 is mixed for pediatrics.
Because our results were unexpected and discordant with our hypothesis and adult data, we suggest caution in extrapolating pharmacogenetic data for SSRIs from adults to children and recommend further studies. The mechanism for the observed increased incidence of sertraline and (es)citalopram AEs in children with normal or increased CYP2C19 metabolism is unknown. Although authors of previous studies have suggested that the ontogeny of CYP2C19 does not change beyond age 1, these authors have not included the increased-function (*17) allele included in this study.19,20 It is important to note that our cohort was a primarily adolescent population. It is possible that there is a pediatric- or adolescent-specific interaction of sertraline or (es)citalopram AEs and CYP2C19. This interaction could be mediated by a specific metabolite in those with normal or increased CYP2C19 metabolism related to the *17 allele that increases the risk for AEs. It is also possible that there is altered expression of cytochrome P450 enzymes during adolescence that affects the sertraline and (es)citalopram metabolism pathway. Our cohorts were predominantly female; there is evidence that estrogen inhibits CYP2C19 expression.21 Although sex was not significant in multivariable analysis, it is possible that the hormonal physiologic changes of pubertal development alter CYP2C19 expression uniquely in female adolescents. There may also be unique pharmacodynamic effects in adolescents.
Although the mechanism for the discordance of our findings to our hypothesis remains unclear, there is previous pediatric literature that supports pediatric-specific effects. For example, Wagner22 demonstrated pediatric-specific directions of effect for drug-gene interactions in certain races. In addition, pediatric population pharmacokinetic models for pantoprazole, another CYP2C19 substrate, reveal large variability despite including CYP2C19 genotype, suggesting variability in CYP2C19 function among those with the same genotype.23
Univariate analysis also revealed fewer Hispanic PMs and IMs compared with NMs for sertraline but not (es)citalopram. Although ethnicity was not significant when included as a covariate in multivariable analysis, further investigation of this association in a larger cohort is warranted. To our knowledge, there is no previous literature on the association of AE reporting by ethnicity.
There are several limitations to this study. In this study, we used a repository of DNA at VUMC that required a previous blood draw and thus may not reflect all pediatric patients on sertraline and (es)citalopram. Among those identified as (es)citalopram exposed, those meeting the inclusion criteria were younger than those excluded, which may also affect generalizability. AEs were identified retrospectively in this observational study by using the EHR for a variety of providers across many departments. Thus, determination of AE status is likely limited by incomplete AE data due to insufficient or inaccurate documentation as well as under- or overidentification of an AE, depending on the provider. There was no causality assessment performed for SSRI exposure and AEs. For many individuals in the study, there was no way to identify medication adherence because it was often not documented and other sources to identify adherence are not available.
Conclusions
Our observations of the relationship of CYP2C19 metabolizer status and AEs in pediatric patients exposed to sertraline or (es)citalopram are discordant with adult data. Our results unexpectedly reveal that children with normal CYP2C19 activity have increased sertraline AEs compared to PMs and IMs. This is not seen in the primary analysis of (es)citalopram, although secondary analysis reveals increased AE risk in those with increased metabolism. This contrasts with adult data, which suggests increased sertraline and (es)citalopram AEs in individuals with reduced CYP2C19 metabolizer status. Considering the retrospective nature of this study and lack of causality assessment with this design, we recognize the limitations of this study and do not recommend changing pediatric prescribing practices of sertraline and (es)citalopram based on CYP2C19 function at this time. However, these unexpected findings highlight the need for further analysis of sertraline and (es)citalopram AEs in larger pediatric cohorts to determine the relevance of CYP2C19 variants in guiding therapy for this population. Clinicians should use caution when using adult-based pharmacogenetic guidelines to inform medication prescription in the pediatric population, particularly for sertraline or (es)citalopram and CYP2C19.24
Glossary
- AE
adverse event
- CI
confidence interval
- CPIC
Clinical Pharmacogenetics Implementation Consortium
- CYP2C19
cytochrome P450 2C19
- EHR
electronic health record
- (es)citalopram
escitalopram and citalopram
- HR
hazard ratio
- IM
intermediate metabolizer
- IQR
interquartile range
- NM
normal metabolizer
- PM
poor metabolizer
- RM
rapid metabolizer
- SSRI
selective serotonin reuptake inhibitor
- TBD
to be determined
- UM
ultrarapid metabolizer
- VUMC
Vanderbilt University Medical Center
Footnotes
Dr Rossow and Mrs Aka conceptualized and designed the study, acquired the data, analyzed and interpreted the data, and drafted the initial manuscript; Dr Maxwell-Horn analyzed and interpreted the data; Dr Roden conceptualized and designed the study; Dr Van Driest conceptualized and designed the study and analyzed and interpreted the data; and all authors revised the manuscript critically for important intellectual content, approved the final manuscript as submitted, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
FINANCIAL DISCLOSURE: Dr Van Driest has been an invited speaker to Merck; the other authors have indicated they have no financial relationships relevant to this article to disclose.
FUNDING: This work used a data set from Vanderbilt University Medical Center’s BioVU and was stored in Research Electronic Data Capture, which are supported by institutional funding and by the Clinical and Translational Science Award UL1 TR000445 from the National Institutes of Health’s National Center for Advancing Translational Sciences. Dr Van Driest was also supported by Burroughs Wellcome Fund Innovation in Regulatory Science award 1015006 and Dr Rossow by the National Institutes of Health’s National Institute of General Medical Sciences Clinical Pharmacology Training Program 5T32 GM007569. Funded by the National Institutes of Health (NIH).
POTENTIAL CONFLICT OF INTEREST: The authors have indicated they have no potential conflicts of interest to disclose.
References
- 1.Lochmann D, Richardson T. Selective serotonin reuptake inhibitors. Handb Exp Pharmacol. 2019;250:135–144 [DOI] [PubMed] [Google Scholar]
- 2.Olfson M, He J-P, Merikangas KR. Psychotropic medication treatment of adolescents: results from the National Comorbidity Survey-Adolescent Supplement. J Am Acad Child Adolesc Psychiatry. 2013;52(4):378–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rynn MA, Walkup JT, Compton SN, et al. . Child/adolescent anxiety multimodal study: evaluating safety. J Am Acad Child Adolesc Psychiatry. 2015;54(3):180–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.US National Library of Medicine. Sertraline hydrochloride - sertraline hydrochloride tablet. 2008. Available at: https://dailymed.nlm.nih.gov/dailymed/drugInfo.cfm?setid=a8b2ad71-cfdc-4a29-8194-deefea138b16&audience=consumer. Accessed June 2, 2019
- 5.Cipriani A, Zhou X, Del Giovane C, et al. . Comparative efficacy and tolerability of antidepressants for major depressive disorder in children and adolescents: a network meta-analysis. Lancet. 2016;388(10047):881–890 [DOI] [PubMed] [Google Scholar]
- 6.Hicks JK, Bishop JR, Sangkuhl K, et al.; Clinical Pharmacogenetics Implementation Consortium . Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther. 2015;98(2):127–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowton E, Field JR, Wang S, et al. . Biobanks and electronic medical records: enabling cost-effective research. Sci Transl Med. 2014;6(234):234cm3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roden DM, Pulley JM, Basford MA, et al. . Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGregor TL, Van Driest SL, Brothers KB, Bowton EA, Muglia LJ, Roden DM. Inclusion of pediatric samples in an opt-out biorepository linking DNA to de-identified medical records: pediatric BioVU. Clin Pharmacol Ther. 2013;93(2):204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research Electronic Data Capture (REDCap)–a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Indiana University School of Medicine Drug interactions Flockhart table: cytochrome P450 drug interaction table. 2007. Available at: https://drug-interactions.medicine.iu.edu/Main-Table.aspx. Accessed June 1, 2020
- 12.Caudle KE, Dunnenberger HM, Freimuth RR, et al. . Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med. 2017;19(2):215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pharmacogene Variation consortium. CYP2C19. Available at: https://www.pharmvar.org/gene/CYP2C19. Accessed August 5, 2019
- 14.Swen JJ, Nijenhuis M, de Boer A, et al. . Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89(5):662–673 [DOI] [PubMed] [Google Scholar]
- 15.Poweleit EA, Aldrich SL, Martin LJ, Hahn D, Strawn JR, Ramsey LB. Pharmacogenetics of sertraline tolerability and response in pediatric anxiety and depressive disorders. J Child Adolesc Psychopharmacol. 2019;29(5):348–361 [DOI] [PubMed] [Google Scholar]
- 16.AlOlaby RR, Sweha SR, Silva M, et al. . Molecular biomarkers predictive of sertraline treatment response in young children with fragile X syndrome. Brain Dev. 2017;39(6):483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aldrich SL, Poweleit EA, Prows CA, Martin LJ, Strawn JR, Ramsey LB. Influence of CYP2C19 metabolizer status on escitalopram/citalopram tolerability and response in youth with anxiety and depressive disorders. Front Pharmacol. 2019;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlsson B, Olsson G, Reis M, et al. . Enantioselective analysis of citalopram and metabolites in adolescents. Ther Drug Monit. 2001;23(6):658–664 [DOI] [PubMed] [Google Scholar]
- 19.Sanford JC, Guo Y, Sadee W, Wang D. Regulatory polymorphisms in CYP2C19 affecting hepatic expression. Drug Metabol Drug Interact. 2013;28(1):23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koukouritaki SB, Manro JR, Marsh SA, et al. . Developmental expression of human hepatic CYP2C9 and CYP2C19. J Pharmacol Exp Ther. 2004;308(3):965–974 [DOI] [PubMed] [Google Scholar]
- 21.Mwinyi J, Cavaco I, Pedersen RS, et al. . Regulation of CYP2C19 expression by estrogen receptor α: implications for estrogen-dependent inhibition of drug metabolism. Mol Pharmacol. 2010;78(5):886–894 [DOI] [PubMed] [Google Scholar]
- 22.Wagner JB. Children are not small adults: specific findings in statin exposure and response in a growing population. Clin Pharmacol Ther. 2019;106(2):278–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shakhnovich V, Brian Smith P, Guptill JT, et al.; Best Pharmaceuticals for Children Act–Pediatric Trials Network . A population-based pharmacokinetic model approach to pantoprazole dosing for obese children and adolescents. Paediatr Drugs. 2018;20(5):483–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lesche D, Mostafa S, Everall I, Pantelis C, Bousman CA. Impact of CYP1A2, CYP2C19, and CYP2D6 genotype- and phenoconversion-predicted enzyme activity on clozapine exposure and symptom severity. Pharmacogenomics J. 2020;20(2):192–201 [DOI] [PubMed] [Google Scholar]