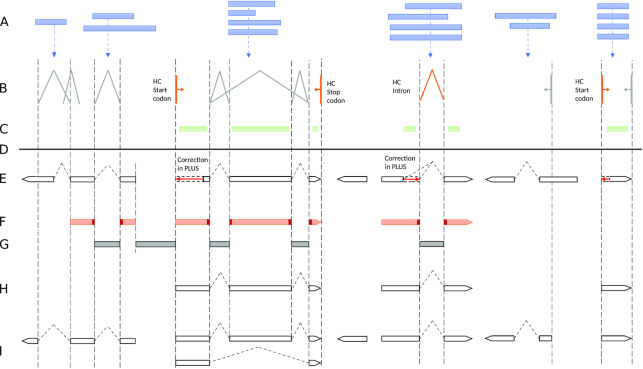
Figure 2.
Evidence integration in BRAKER2. (A) Target proteins; (B) Introns, gene start and stop sites defined by spliced alignments of target proteins to genome; (C) CDSpart chains; (D) Genome sequence; (E) Genes predicted by GeneMark-EP+ at a given iteration. The high confidence hints are enforced (red arrows); (F) Anchored sites, the splice sites and gene ends predicted ab initio and corroborated by protein hints; (G) Anchored introns and intergenic sequences bounded by anchored gene ends are selected into training of non-coding sequence model for GeneMark-EP+; (H) Anchored multi-exon and single exon genes predicted by GeneMark-EP+ and selected for training AUGUSTUS; (I) Transcripts predicted by AUGUSTUS with support of an external evidence.