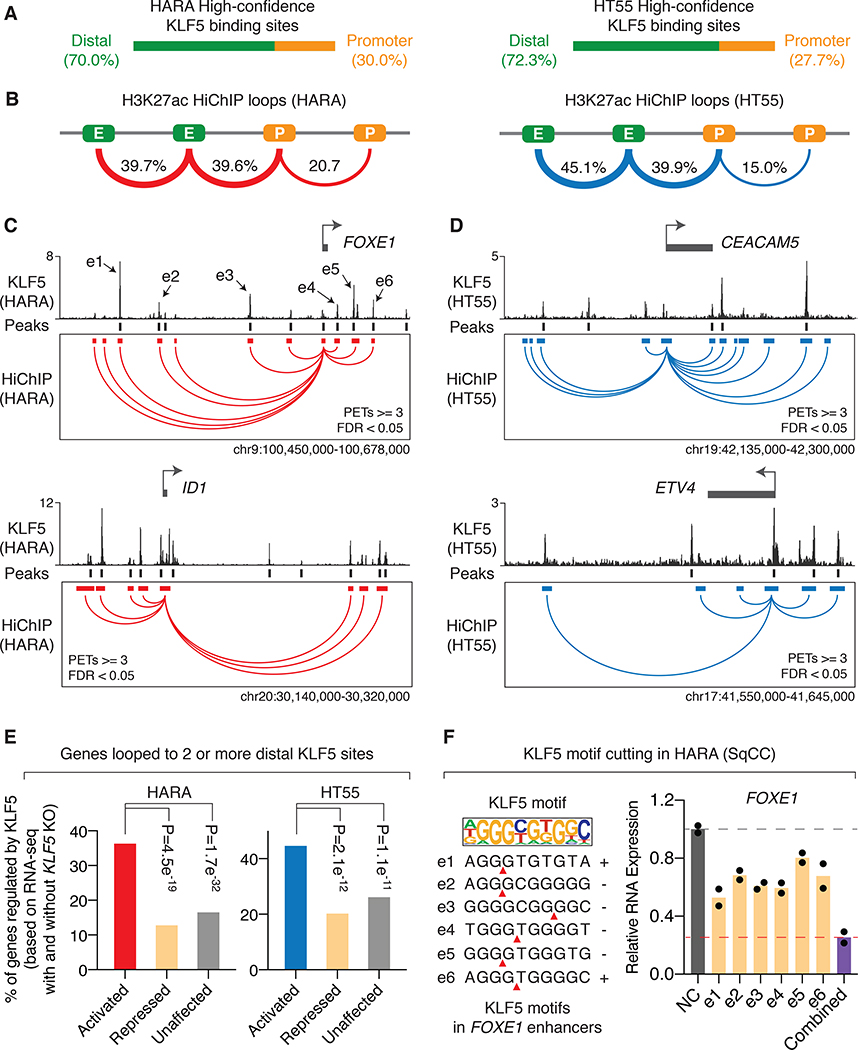
Figure 3: Chromatin loops aggregate distal KLF5 binding events to activate their target genes.
A. Distribution of high-confidence KLF5 binding sites (KLF5 binding sites that were called in biological replicates of ChIP-seq and harboring KLF5 DNA binding motifs) in HARA and HT55 cells.
B. Distribution of significant chromatin loops called from H3K27ac HiChIP assays (PETs≥3, FDR<0.05) in HARA and HT55 cells. E: enhancers; P: promoters.
C. KLF5-activated genes FOXE1 and ID1 (based on RNA-seq results in HARA cells with and without KLF5 knockout, FDR<0.05) are connected to multiple high-confidence distal KLF5 binding sites through chromatin loops identified from H3K27ac HiChIP assays in HARA squamous cancer cells. HiChIP anchors that are looped to promoters of the target genes are presented.
D. Same as C, but with regards to KLF5-activated genes CEACAM5 and ETV4 in HT55 colorectal cancer cells.
E. Percentage of KLF5-activated, -repressed, and -unaffected genes (based on RNA-seq results with and without KLF5 knockout, FDR<0.05) that are connected to two or more high-confidence distal KLF5 binding sites in HARA and HT55 cells. P values are derived from Fisher Exact Tests.
F. Expression level of the KLF5-activated gene FOXE1 after Cas9-mediated individual disruption of KLF5 motifs within each of FOXE1-connected distal KLF5 binding sites, or simultaneous disruption of the motifs. Gene expression level is normalized to the negative control.