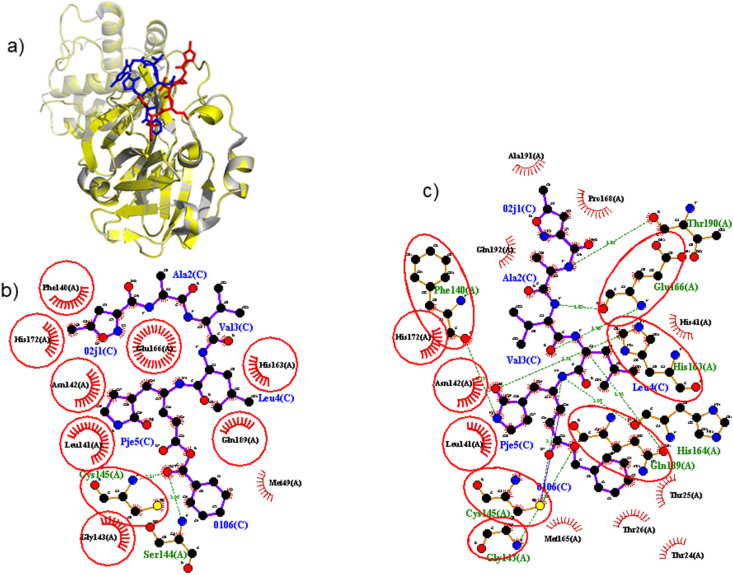
Figure 3.
Molecular docking protocol validation. This crucial process can enhance the accuracy and reliability of an in-silico docking experiment. A comparison of the binding modes for the re-docked ligand (blue) vs. the co-crystallized ligand (red), shown as a stick representation. a) The molecular docking protocol accurately regenerated the binding configuration of a crystallographically determined protein-ligand complex with an RMSD value of 1.049 Å using PyMOL. Amino acid residue interactions with b) the re-docked and c) the co-crystallized ligand executed in LigPlot+.