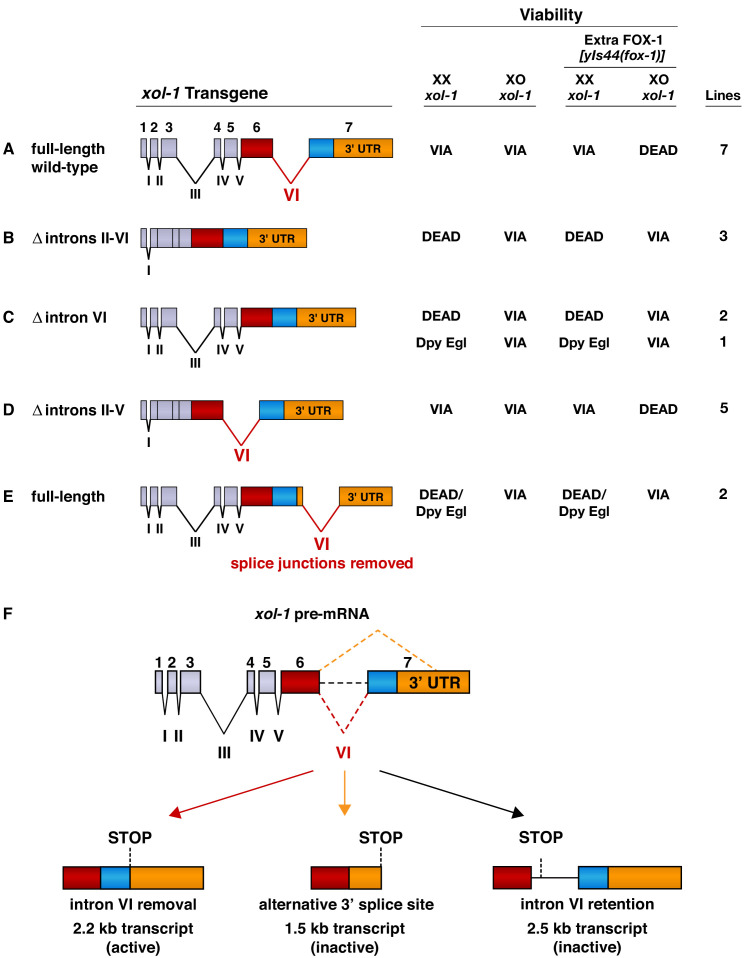
Figure 2. Intron VI is essential for repression of xol-1 by FOX-1.
(A–E) Assays of wild-type xol-1 transgenes and deletion derivatives with different combinations of introns show that removal of intron VI prevents FOX-1 from repressing xol-1. Diagrams on the left show the intron–exon structure of xol-1 sequences in the transgene derivatives. Exon 7 includes both coding sequences (blue) and the 3' UTR (orange). The wild-type parent transgene rescues the XO-specific lethality caused by xol-1 null mutations but permits XX animals to be viable. Intron deletion derivatives have genomic regions of xol-1 replaced by corresponding xol-1 cDNA to remove introns without altering protein coding sequence. Shown on the right is the viability of XX and XO xol-1(y9) deletion mutant animals carrying extra-chromosomal arrays of the different transgene derivatives. The arrays were made and assayed in him-5(e1490); xol-1(y9) mutants that produce 33% XO and 67% XX embryos and then crossed into him-5; xol-1 strains producing high levels of FOX-1 from an integrated array [yIs44 (fox-1)] carrying multiple copies of fox-1. The far right shows the number of independent extra-chromosomal arrays assayed. The term ‘dead’ means that all animals of the genotype were inviable, and no extra-chromosomal arrays could be established in XX animals. The arrays could only be established and maintained through XO males. The term ‘via’ means the animals were viable and appeared wild type. The term ‘Dpy Egl’ refers to the phenotype of dosage-compensation-defective XX animals that escape lethality. XX animals are typically dumpy (Dpy) in body size and egg-laying defective (Egl). The term ‘dead/Dpy Egl’ means that most animals (greater than 90%) were dead, and rare escapers were Dpy Egl. High levels of FOX-1 kill all XO animals only if a spliceable form of intron VI is present. FOX-1 does not repress xol-1 in XO animals if intron VI is absent or is relocated to the 3' UTR without splice junctions. Transgenes lacking intron VI kill XX animals, as do wild-type xol-1 transgenes in strains with a fox-1 null mutation, because all transgenes are expressed at a somewhat higher level than the endogenous xol-1 gene in XX animals. (F) Structures of the three most abundant splice variants of xol-1 transcripts are shown. Only the 2.2 kb variant, which lacks intron VI (red dashed line) and includes essential exon 7 coding sequences (blue) and 3' UTR, is necessary and sufficient for survival of XO animals. This isoform corresponds to Wormbase.org transcript C18A11.5b.1. Transcripts that retain intron VI (2.5 kb isoform corresponding to Wormbase.org transcript C18A11.5c.1) (black dashed line) or lack exon 7 coding sequences (1.5 kb isoform corresponding to Wormbase.org transcript C18A11.5a) (orange dashed line) due to use of an alternative 3' splice acceptor site cannot produce essential XOL-1 male-determining proteins.