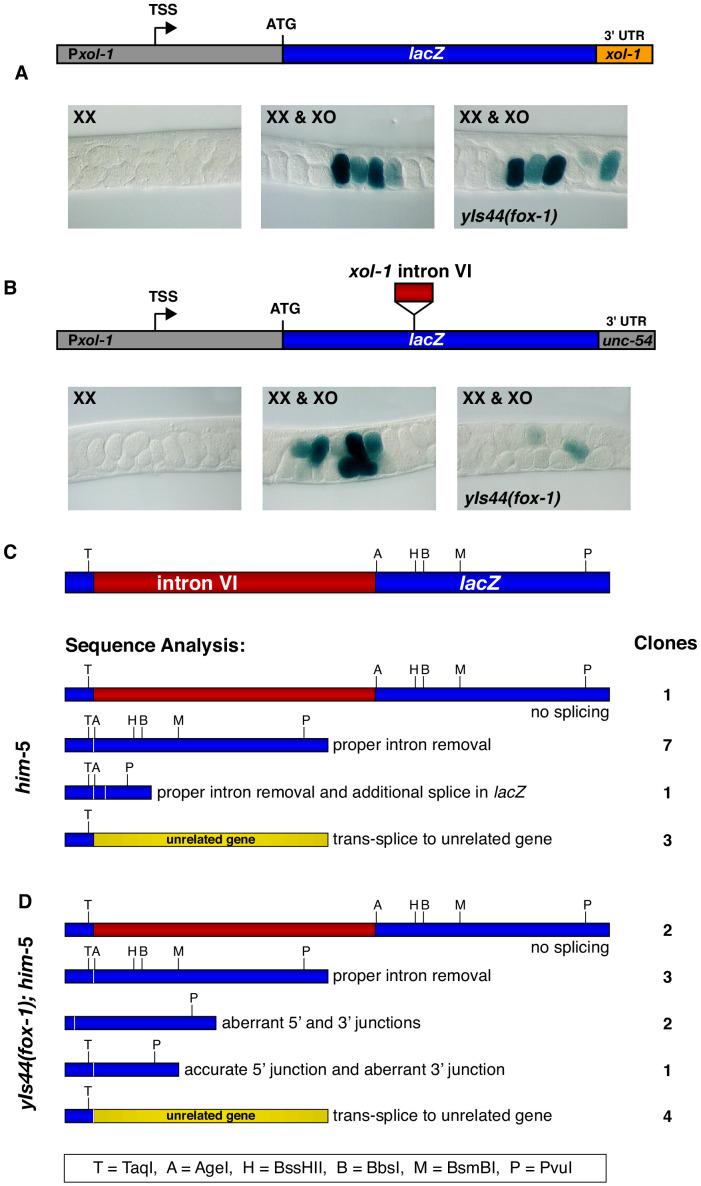
Figure 4. Intron VI of xol-1 is sufficient to confer FOX-1 repression.
(A) The promoter and 3' UTR of xol-1 are not sufficient for FOX-1 to repress xol-1. Below the diagram of the Pxol-1::lacZ::xol-1 3' UTR reporter transgene (pMN27) are sections of adult gonads from different genotypes stained with 5-bromo-4-chloro-3-indolyl-D-galactopyranoside. Genotypes of embryos in the gonads include: (left) XX, unc-76; yEx231 [pMN27 and unc-76 (+)]; (middle) XX and XO, him-5 unc-76; yEx231 [pMN27 and unc-76 (+)]; (right) XX and XO, yIs44(fox-1); him-5 unc-76; yEx231 [pMN27 and unc-76 (+)]. The lacZ reporter is sex-specifically regulated: high levels of β-galactosidase in XO embryos but low levels in XX embryos. High levels of FOX-1 do not diminish β-galactosidase activity in the absence of intron VI, indicating that the xol-1 promoter and xol-1 3' UTR cannot confer FOX-1 repression. Five independent extra-chromosomal array strains of each genotype carrying pMN27 showed the results represented. At least 1000 embryos were examined for each genotype derived from each of the five independent arrays. (B) Intron VI of xol-1 is sufficient for FOX-1 to repress a lacZ reporter gene. Shown is a diagram of the Pxol-1::lacZ::intronVI::unc-54 3' UTR reporter transgene (pMN110) in which the 3' UTR is from the body-wall myosin gene unc-54. Genotypes of adult gonads stained for β-galactosidase activity are the same as listed in (A), except the array is yEx280 [(pMN110) and unc-76 (+)]. This intron VI-containing lacZ reporter is also sex-specifically regulated: active in XO embryos and repressed in XX embryos. High levels of FOX-1 (from yIs44) greatly diminish the level of β-galactosidase activity in XO embryos, indicating that intron VI alone is sufficient for FOX-1 repression. Four independent extra-chromosomal array strains of each genotype carrying pMN110 showed the results represented. At least 1000 embryos were examined for each genotype derived from each of the four independent arrays. (C, D) Sequence analysis of cDNA clones from lacZ transcripts shows that excess FOX-1 increases intron VI retention and also causes alternative pre-mRNA splicing using 3' splice acceptor sites in lacZ via cis-splicing and also 3' splice acceptor sites in unrelated genes via trans-splicing. Below the diagram of lacZ's relevant intron–exon structure and restriction sites is the sequence analysis of lacZ cDNAs from him-5 (C) and from yIs44(fox-1); him-5 (D) strains. Shown are the splicing patterns revealed by DNA sequence analysis and also the number of lacZ clones with each indicated structure. During trans-splicing, the proper 5' donor at the lacZ exon–intron VI junction was used in combination with a naturally occurring 3' acceptor at an intron–exon junction of an unrelated gene (see text).